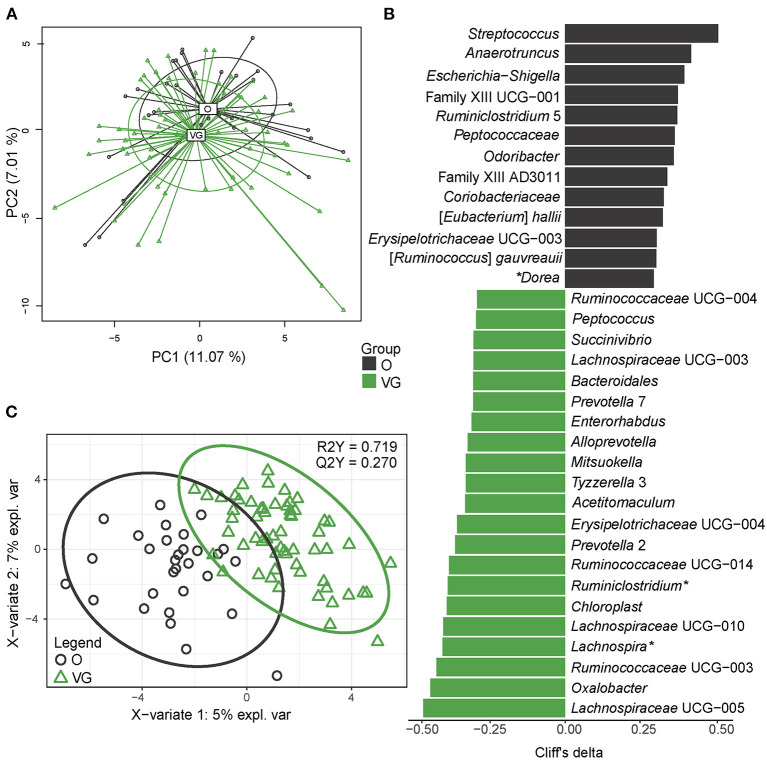
Figure 1.
Gut microbiome composition. (A) Two-dimensional (2D) principal component analysis (PCA) score plot with the explained variance of each component. (B) Biomarker taxa generated from univariable discrimination analysis (FDR ≥ 0.1), effect size estimated by Cliff's delta; (C) 2D score plots of PLS-DA. R2Y fit goodness, Q2Y predictive power. *genera belonging to the core microbiome (abundance > 0.1%, prevalence > 75%). Effect sizes, FDR values, and variable importance in projection (VIP) values can be found in Supplementary Table 3. Data are presented as compositional (proportion of the particular bacteria of total sum of bacteria) after clr transformation.