FIGURE 4.
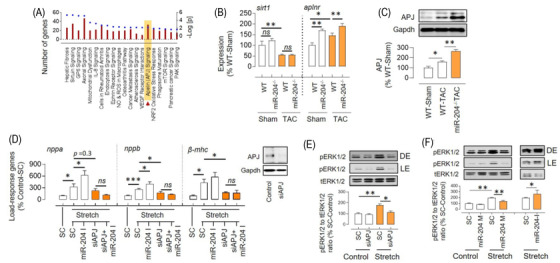
miR‐204 inversely regulates APJ‐mediated stretch response. (A) Top 20 pathways that differ in hearts of WT‐TAC versus miR‐204–/– TAC mice, based on ingenuity pathway analysis of gene expression profiles. The left y‐axis indicates the number of genes linked with the pathway (bar), and the right y‐axis indicates the significance (dot) (n = 4). (B) The expression levels of sirt1 and aplnr genes in the heart of WT and miR‐204−/− sham/TAC mice (n = 5–15). (C) Increased levels of APJ in the heart of miR‐204–/– TAC mice compared to that of WT‐TAC mice (n = 5). (D) Effect of APJ silencing (50 nM) and miR‐204 inhibition (100 nM) on load‐response gene expression in H9C2 cells during the stretch (n = 6). Insert shows effect of siRNA (50 nM) on APJ silencing. (E) The stretch‐induced ERK1/2 activation in the setting of APJ silencing (50 nM) (n = 3). (F) Effect of miR‐204 mimic (M, 100 nM) and inhibitor (I, 100 nM) on the stretch‐induced increase in pERK1/2 in H9C2 cells. Data are shown as mean, and error bars represent SEM (n = 4). *p < .05, **p < .01, versus indicated group. SC, scrambled control; pERK1/2, phosphorylated extracellular signal‐regulated kinase 1/2; tERK1/2, total extracellular signal‐regulated kinase 1/2; nppa, natriuretic peptide a; nppb, natriuretic peptide b; β‐mhc, beta‐myosin heavy chain; LE, light exposure; DE, dark exposure
