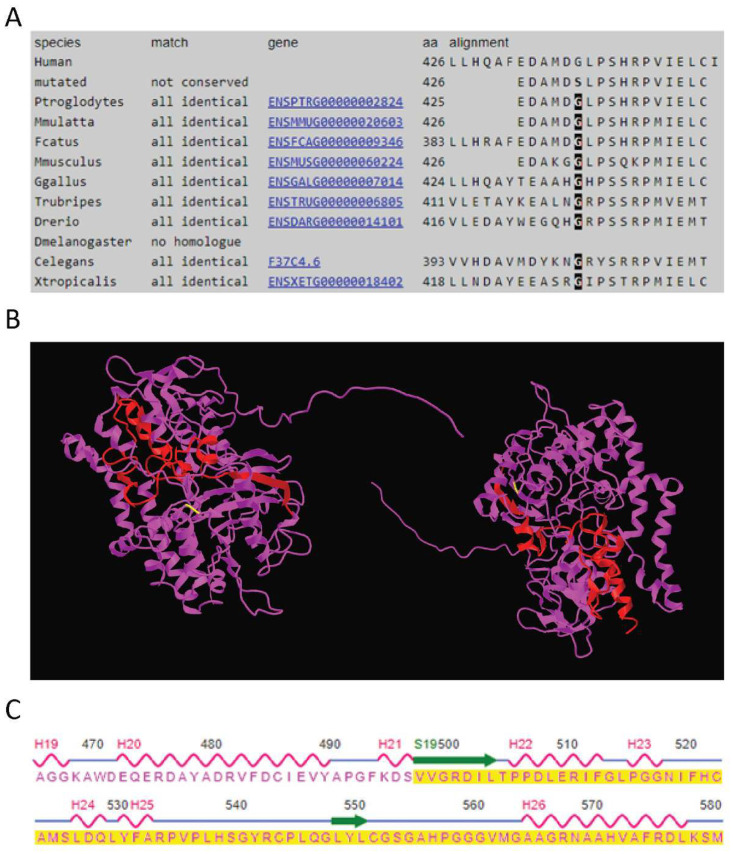
Figure 3.
Evolutionary sequence conservation and 3D structure of PYROXD2. (A) The multiple sequence alignment of PYROXD2 reveals the affected p.Gly426 residue is highly conserved. (B) The predicted 3D structure of wild-type PYROXD2 (AF-Q8N2H3-F1, AlphaFold, modelled in icn3d) highlighting the patient variant p.(Gly426Ser) is highlighted in yellow, and the wild-type residues affected by the frameshift variant p.(Val498Cysfs*79) are highlighted in red. (C) Structural regions (from icn3d) indicate that the frameshift variant starting at p.Val498 (region highlighted in yellow) affects a beta-fold region (amino acids 498–504) and two major alpha-helical regions (amino acids 506–513 and 565–578) of the PYROXD2 protein.