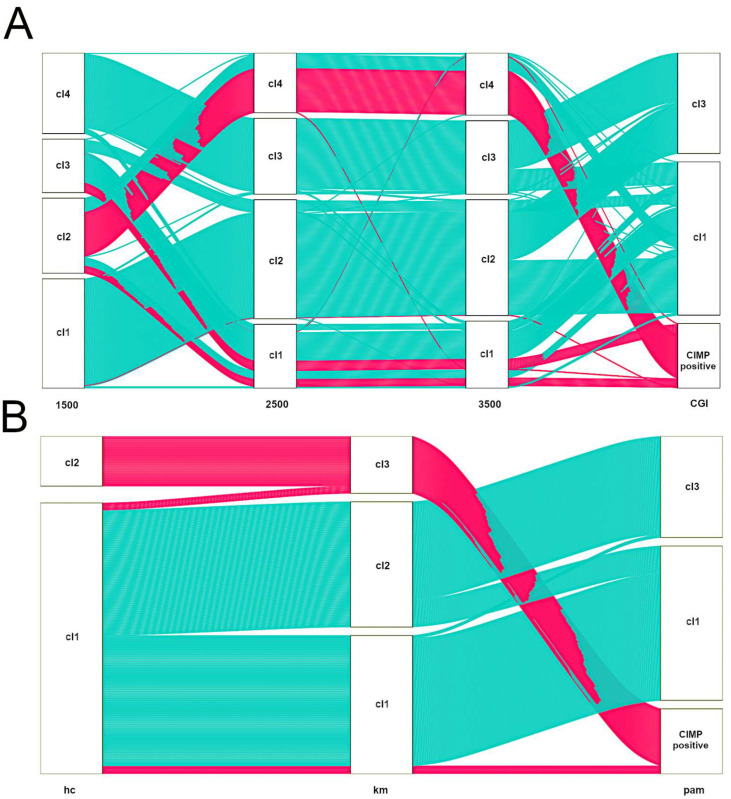
Figure 3.
Alluvial diagrams illustrating the influence of choice of variable selection strategy (A) or clustering algorithm (B) on CIMP-positive assignment in colon cancer (TCGA dataset). In all cases, we used the consensus clustering approach with Euclidean distance and average linkage [120]. Consensus clustering was run using 80% sample resampling, a maximum evaluated k of 8 and 1000 resamplings. (A). Samples were clustered by pam (partitioning around medoids). All together, 1500 or 2500 or 3500 most variable Illumina 450K probes or probes located in CGI that displayed cancer-specific methylation were selected. CIMP-positive tumours identified by the last approach are represented with pink alluvia. Note changes in the flow of CIMP-positive samples from left to right. (B) For clustering, probes located in CGI that displayed cancer-specific methylation were selected. Samples were clustered by selecting hierarchical clustering (hc) or k-means (km) or partitioning around medoids (pam). CIMP-positive tumours identified by the last approach are represented with pink alluvia. Note changes in the flow of CIMP-positive samples from left to right.