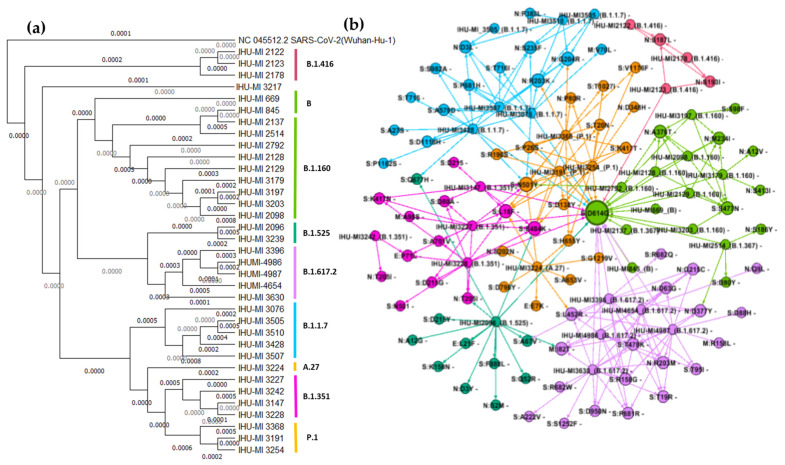
Figure 3.
Genetic diversity of the SARS-CoV-2 isolates used. (a) Phylogenetic analysis of isolated SARS-CoV-2 presented through a maximum likelihood tree, with a bootstrap analysis based on the values of 1,000 replicates. The isolates were grouped according to the PANGO classification, based on the analysis previously obtained from the Nextclade tool. (b) Distribution of the mutations that generate amino acid exchange in structural proteins of SARS-CoV-2 among the isolates used in the study. Arrows link the isolates to their identified mutations; the size of the nodes represents the gradient incoming (the most shared mutations are in bigger circles). Legend: E, envelope protein; M, membrane protein; N, nucleocapsid protein; S, spike protein.