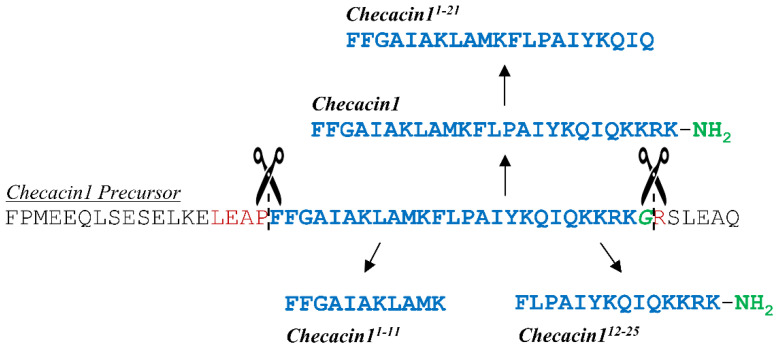
Figure 1.
Precursor sequence of Checacin1 (without signal peptide) and the sequences used for the bioassays. Native Checacin1 is N-terminally cleaved at a highly effective ‘LEAP’ motif [22], which is typical of most checacin precursors [23]. The C-terminal cleavage of the Checacin1 progenitor is upstream of a monobasic (Arg) cleavage site and has a C-terminal amidation site (Gly). The preceding KKRK motif does not function as a cleavage signal [22]. Checacin11−11 and Checacin112−25 are naturally occurring fragments of Checacin1, while Checacin11−21 has not been detected in the venom of C. cancroides [22]. However, for the orthologous Megicin 18 from the scorpion M. gibbosus, the C-terminal cleavage was postulated to be upstream of such a tetrabasic motif [24].