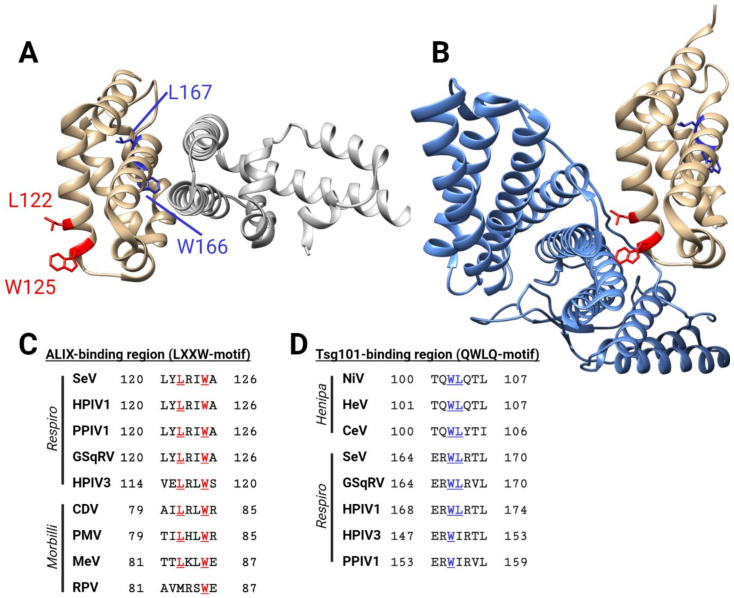
Figure 3.
Structural features of the SeV C protein. (A) SeV C protein (amino acid residues 99–204, shown in gold) in a complex with STAT1 (silver) (PDB 3WWT) [59]. Residues important for binding to ALIX (LXXW motif) are shown in red; potential Tsg101-binding residues (QWLQ-motif) are shown in blue. (B) SeV C protein (amino acid residues 99–204, shown in gold) in a complex with the BRO1 domain of ALIX (light blue) (PDB 6KP3) [61]. (C) Conserved ALIX-binding motifs in respiroviruses (top five sequences), and homologous residues in morbilliviruses (bottom four sequences). (D) Conserved Tsg101-binding motifs in henipaviruses (top three sequences), and homologous residues in respiroviruses (bottom 5 sequences). Figure generated with BioRender.com.