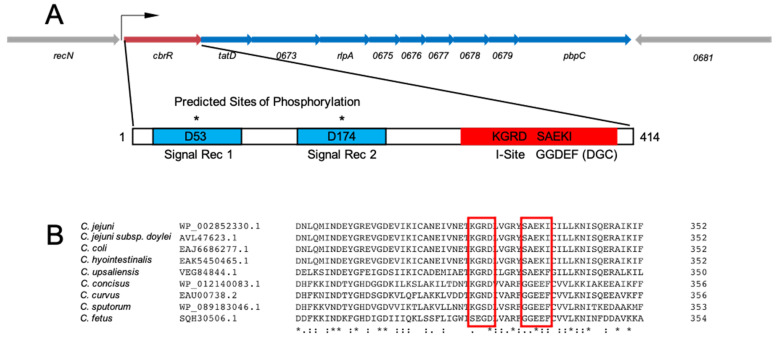
Figure 1.
Gene and protein schematics for CbrR and the cbrR operon in C. jejuni 81–176. (A) The cbrR gene is the first of a predicted ten gene operon. For clarity, locus tags have been abbreviated to the last four digits. The bent arrow indicates the predicted transcription start site [68]. CbrR is 414 amino acid residues in length and is comprised of two N-terminal signal receiver domains (blue) with predicted sites of phosphorylation D53 and D174, and the GGDEF domain (red), which includes an autoinhibitory site (I-site). The GGDEF domain is indicative of diguanylate cyclase (DGC) activity. (B) A multiple sequence alignment shows that, though non-canonical, the amino acids comprising the I-site and active site are highly conserved within several clinically relevant Campylobacter species. Amino acid conservation is indicated by either an * (asterisk; fully conserved residues); a : (colon; amino acids with strongly similar properties; or a . (period; amino acids with weakly similar properties).