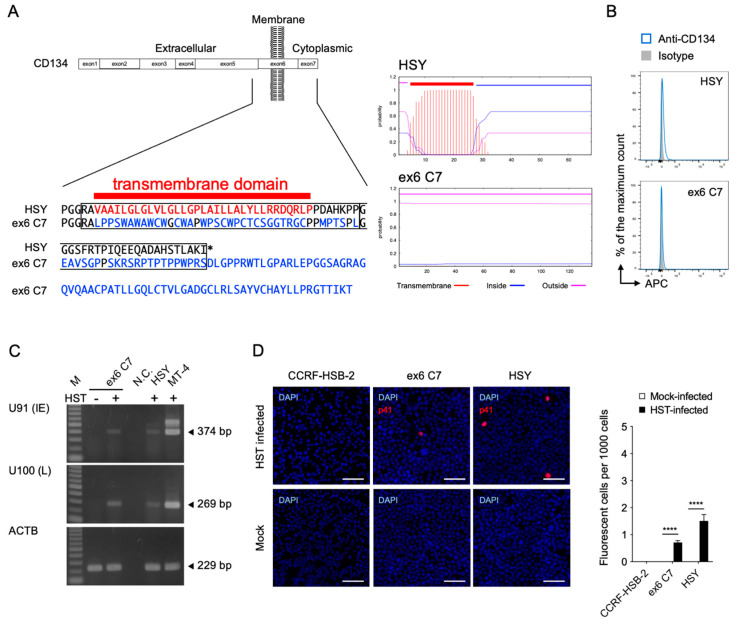
Figure 2.
Establishment of TNFRSF4 gene-specific mutated HSY cells (ex6 C7 cells) and HHV-6B permissivity. (A) Comparison of amino acid sequences and predicted transmembrane helices around the target region in TMHMM2.0 [41]. In ex6 C7 cells, a frameshift mutation near the N-terminus of the CD134 transmembrane region (left panel) led to translation of an amino acid sequence not capable of serving as a transmembrane region (right panels). (B) CD134 expression by HSY cells and ex6 C7 cells was analyzed by means of fluorescence-activated cell sorting analysis. (C) Viral RNA detection in ex6 C7 cells by RT-PCR. Cells were infected with HHV-6B HST and total RNA was extracted at 48 h post-infection. U91 (immediate early) and U100 (late) transcripts were detected. β-actin (ACTB) was used as an internal control. M, DNA ladder; -, mock-infected cells; +, HST-infected cells; N.C., no template control. (D) Viral antigen detection in ex6 C7 cells and HSY cells using immunofluorescence assay. Cells were infected with HHV-6B HST and treated with cold acetone at 48 h post-infection (left panel). HHV-6 p41 protein (red) was detected in both cell types. Nuclei of cells were stained with DAPI (blue). Scale bars represent 50 μm. Virus-infected cells (red) were counted (right panel). Error bars represent standard deviations (n = 3 per cell). Asterisks indicate statistical significance (**** p < 0.0001, Bonferroni’s multiple comparison test).