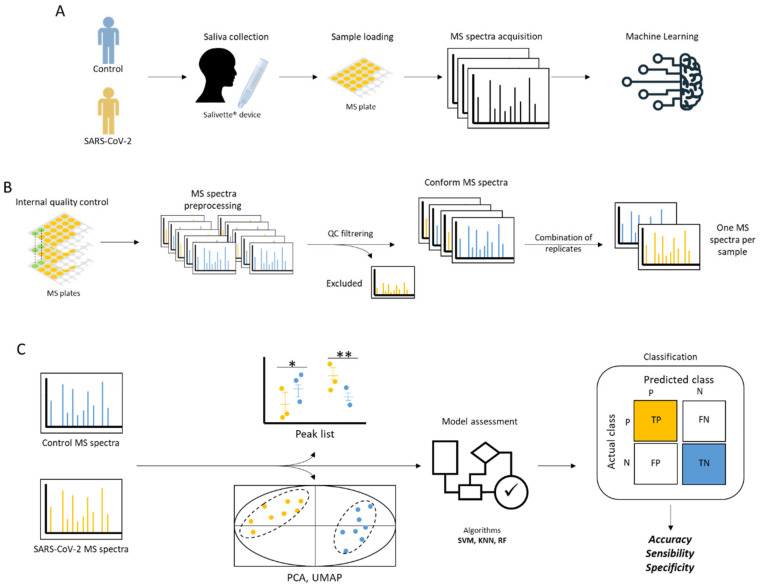
Figure 1.
Schematic presentation of the experimental workflow. (A) Main steps of the study. Individuals who were SARS-CoV-2-positive and negative (controls) based on NPS RT-qPCR tests were enrolled. Saliva was collected with Salivette® devices (SARSTEDT, Numbrecht, Germany), loaded onto an MS plate, and then submitted to MALDI-TOF MS acquisition. The resulting MS spectra were analyzed using machine learning (ML) methods. (B) Pre-processing steps of the MS spectra included a quality control step, evaluated the homogeneity of the data among the Cov+ and Cov− groups, and determined peak detection conditions. (C) Strategy used for the prediction of SARS-CoV-2-positive saliva using ML. Statistical analyses were performed to reveal relevant MS peaks before the assessment of ML models.