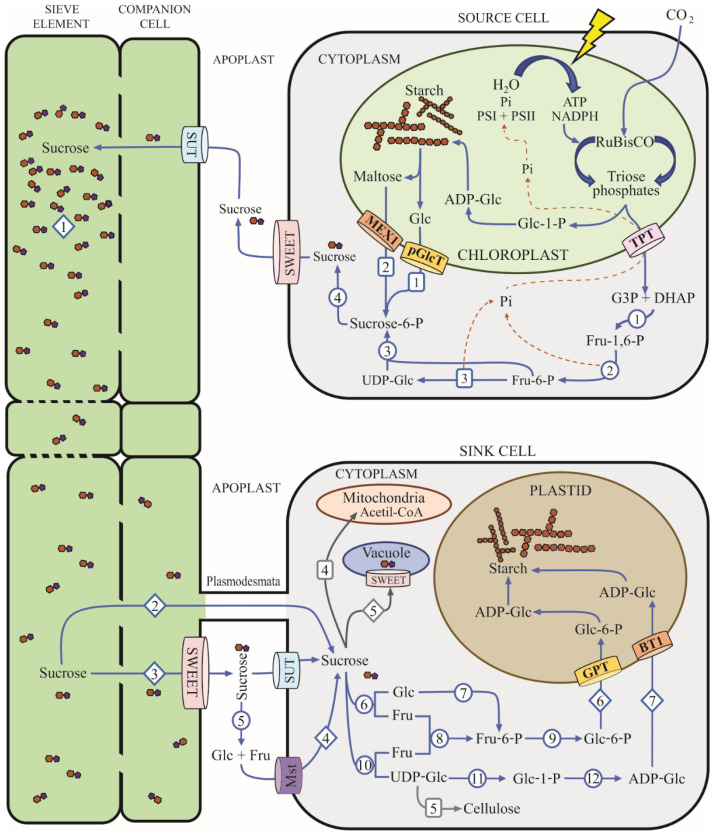
Figure 1.
Common plant pathways of carbon assimilation and partitioning from source to sink tissues. This diagram shows a current model for organic carbon biosynthesis and translocation from photosynthetic to heterotrophic cells (based on previous models by MacNeill et al., 2017 and López-González et al., 2019 [51,52]). Blue arrows trace the current carbon flow routes, the discontinued orange arrows show Pi flux, and the colored barrels designate carbohydrate transporters. Enzymes are indicated in numbered circles as: (1) Fructose 1,6-bisphosphate aldolase (EC 4.1.2.13); (2) FBPase (EC 3.1.3.11); (3) SPS (EC 2.4.1.14); (4) SPP (EC 3.1.3.24); (5) Apoplastic invertase (EC 3.2.1.26); (6) Neutral (cytoplasmic) invertase; (7) Hexokinase (EC 2.7.1.1); (8) Fructokinase (EC 2.7.1.4); (9) Phosphoglucoisomerase (EC 5.3.1.9); (10) SuSy (EC 2.4.1.13); (11) UDPase (EC 2.7.7.9); (12) ADP-glucose pyrophosphorylase (EC 2.7.7.27) [52]. The numbers in rounded rectangles denote specific metabolic pathways: (1) and (2) starch-derived carbohydrates translocated to cytoplasm by Maltose Excess Protein (MEX1; ([52,53])) or Plastidic Glucose Translocator (pGlcT; [52,54]), as possible precursors for Suc biosynthesis; (3) synthesis of UDP-Glucose from Fructose-6-P; (4) Suc catabolism for energy metabolism; (5) Cellulose biosynthesis. Diamonds indicate potential carbon fluxes as: (1) Suc mass flow from source to sink tissues through the phloem; (2) symplastic transport of Suc from phloem to the sink cells; (3) apoplastic transport of Suc from the phloem to sink cells; (4) Monosaccharides transported from the apoplast to sink cells by Monosaccharide transporters (Mst), as potential substrates for Suc biosynthesis; (5) Vacuole import of Suc for transient storage; 6 and (7) Glc-6-P and ADP-Glc imported into plastids by specific transporters (GPT; Glc-6-P/Pi translocator and BT1; Adenine nucleotide transporter Brittle1, respectively, (see [51,52,54] for review) and directed to starch biosynthesis.