Figure 5.
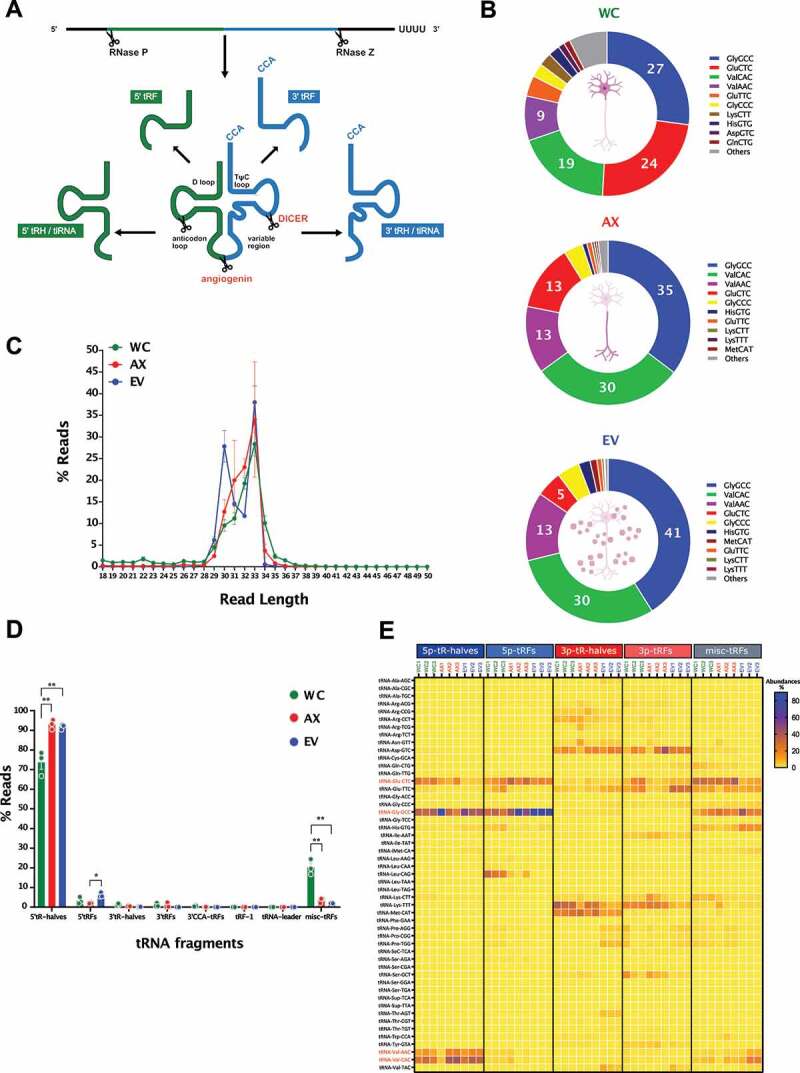
tRNA repertoire in the neuronal subcellular and extracellular compartments. (A) Diagrammatic representation of the biogenesis of tRNA-derived small RNAs (tsRNAs), where mature tRNAs undergo endonuclease cleavage to generate tRNA-derived fragments (3ʹ- and 5ʹ- tRFs) and tRNA halves (3ʹ- and 5ʹ- tRHs). (B) Relative read abundance of parental tRNAs upon tsRNAs mapping in WC, AX and EV shows the most expressed tRNA species in each neuronal compartment (percentage of total tRNA reads). (C) Read length (nt) distribution plot for all tsRNAs (mean±s.e.m.), showing a higher frequency of 33nt long reads in all three of WC, AX and EV samples, but an additional 30nt peak that is predominant in EV samples. (D) Percentage distribution of total reads mapping to each class of tsRNAs in all samples following the unitas annotation workflow (mean ± s.e.m). Comparisons between neuronal compartments demonstrate that 5ʹ-tRHs are the most abundant tsRNAs in all samples and represent a significant higher proportion of AX and EV tsRNAs (~90%) compared to WC (70%). (E) Heatmap of the abundance distribution of tsRNAs present in all WC, AX and EV samples. Data expressed as percentage of total reads per tsRNA class. Highlighted in red are the parental tRNAs generating the most abundant 5ʹ-tRHs: 5ʹ-tRHs-Gly-GCC, 5ʹ-tRHs-Val-AAC, 5ʹ-tRHs-Val-CAC and 5ʹ-tRHs-Glu-CTC. Two-way ANOVA with Tukey’s multiple comparison post-hoc test, * p-value < 0.05; ** p-value < 0.01. Whole Cell (WC), Axon (AX) and Extracellular Vesicles (EV).
