Figure 6.
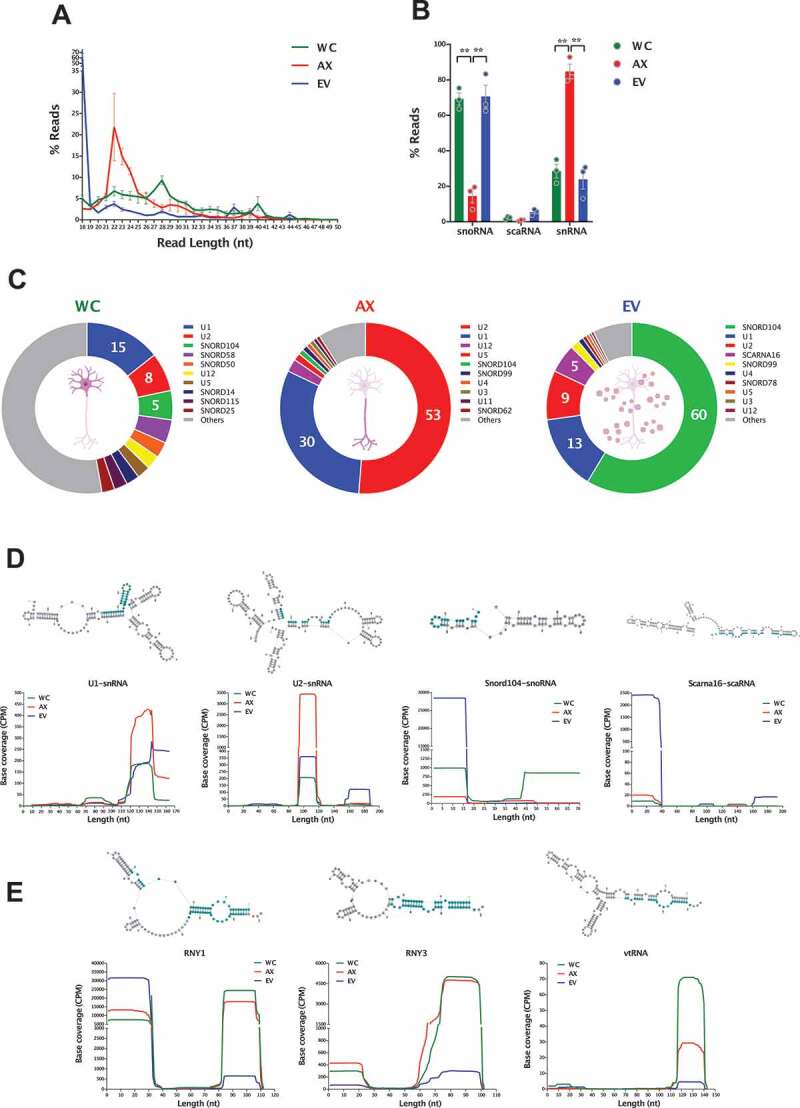
sRNAs (snoRNAs, scaRNAs and snRNAs) in the neuronal subcellular and extracellular compartments. (A) Read length (nt) distribution plot of reads mapping to parental sno/scaRNAs and snRNAs shows the distinct profile between neuronal compartments (mean ± s.e.m.), with a marked 22nt peak in AX samples, which is largely absent in WC and EVs. (B) Mean percentage distribution of total reads mapping to each class of sRNAs investigated in all WC, AX and EV samples (mean ± s.e.m.). Comparisons between neuronal compartments show that snoRNA-derived small RNAs compose the majority of sRNA reads in WC and EV samples (~70%), whereas snRNA fragments are the most abundant class in the AX. (C) Relative abundances of those RNA fragments mapping to parental sno/scaRNAs and snRNAs in WC, AX and EV showing the most expressed in each neuronal compartment. Data expressed as percentage of total sno/scaRNAs and snRNA reads. (D) Schematic secondary structure of consensus sequence (top) and base coverage plots of the most expressed parental sno/scaRNAs and snRNAs (bottom) reveal specific processing into shorter well-defined RNA fragments, whose expression pattern differs in the subcellular and EV compartments. (E) Schematic secondary structure of consensus sequence (top) and base coverage plots of the most expressed parental YRNAs and vtRNAs (bottom) also reveal processing into shorter well-defined RNA fragments. Note how the distinct processing patterns are dependent on subcellular and extracellular localization. Most abundant processed fragments are highlighted in blue in the secondary structures. Two-way ANOVA with Tukey’s multiple comparison post-hoc test, * p-value < 0.05; ** p-value < 0.01. Whole Cell (WC), Axon (AX) and Extracellular Vesicles (EV).
