Figure 7.
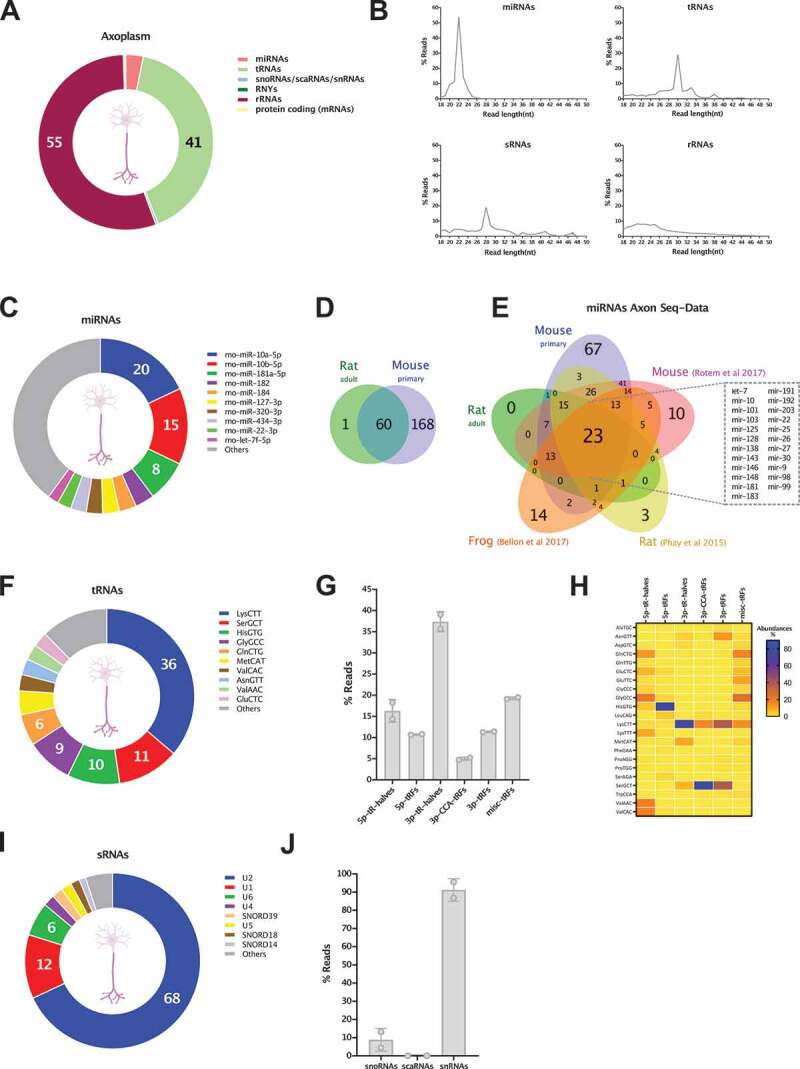
sncRNA profiling of the axoplasm from adult axons. (A) Relative read abundance of each sncRNA biotype in the peripheral nerve’s axoplasm (percentage of total sncRNA reads). (B) Read length (nt) distribution plot of reads mapping to miRNAs, tRNAs, sno/snRNAs (sRNAs) and rRNAs. (C) Relative read abundance of specific miRNAs showing the most expressed in axoplasm samples. Data expressed as percentage of total miRNA reads. (D) Venn diagram illustrates the very high overlap of individual miRNAs detected in the in vivo rat axoplasm (miRNAs ≥ 5 reads) and the in vitro mouse axons (miRNAs ≥ 50 CPM, which is equivalent to ≥ 10 reads). To compare miRNAs of different species only the precursor’s base identification was considered in each case. (E) Venn diagram displaying the overlap of miRNAs detected in our two axon sncRNA-seq datasets (in vivo rat axoplasm and mouse in vitro axons) and the three currently available RNA-seq axon datasets, in vitro rat motor axon, frog retinal ganglion cells axons and rat sciatic nerves. As above, only the precursor’s base identification was considered in each case. This analysis reveals a core of 23 axonal miRNAs present across all neuronal types, both in in vivo and in vitro axons. (F) Relative read abundance of parental tRNAs shows the most expressed parental tRNA species in axoplasm. Data expressed as percentage of total tRNA reads. (G) Percentage distribution of total reads mapping to each class of tRNA-derived small RNA (tsRNAs), following the unitas annotation workflow (mean ± s.e.m). (H) Heatmap of the distribution of each tsRNA class present in the axoplasm with at least 1% of relative abundance. Data expressed as percentage of total reads from tsRNA class. (I) Relative read abundance of parental sno/scaRNAs and snRNAs shows the most expressed in axoplasm samples. Data expressed as percentage of total sno/scaRNAs and snRNAs reads. (J) Percentage distribution of total reads mapping to each class of sRNAs (snoRNAs/scaRNAs/snRNAs) investigated in axoplasm samples (mean ± s.e.m.).
