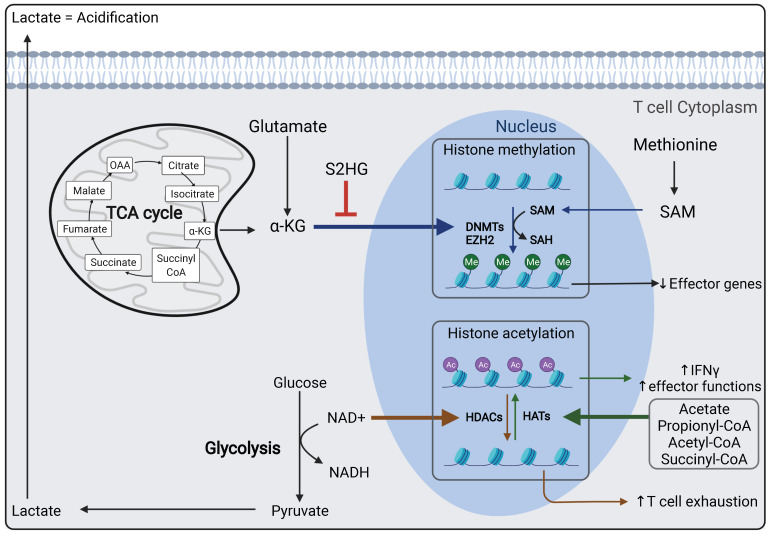
Figure 3.
Immunometabolism shapes the epigenome of T cells. Here several of these epigenetic modifying pathways and metabolites influencing histone modification are outlined. OAA: Oxaloacetate; a-KG: a-Ketoglutarate; S2HG: S-2-hydroxyglutarate; DNMTs: DNA methyltransferase; EZH2: Enhancer of zeste homolog 2; SAM: S-adenosyl methionine; SAH: S-adenosyl homocysteine; NAD+: Nicotinamide adenine dinucleotide; NADH: Nicotinamide adenine dinucleotide hydrogen; HDACs: Histone deacetylases; HATs: Histone acetyltransferase. Figure generated by Biorender.com (accessed on 15 November 2021).