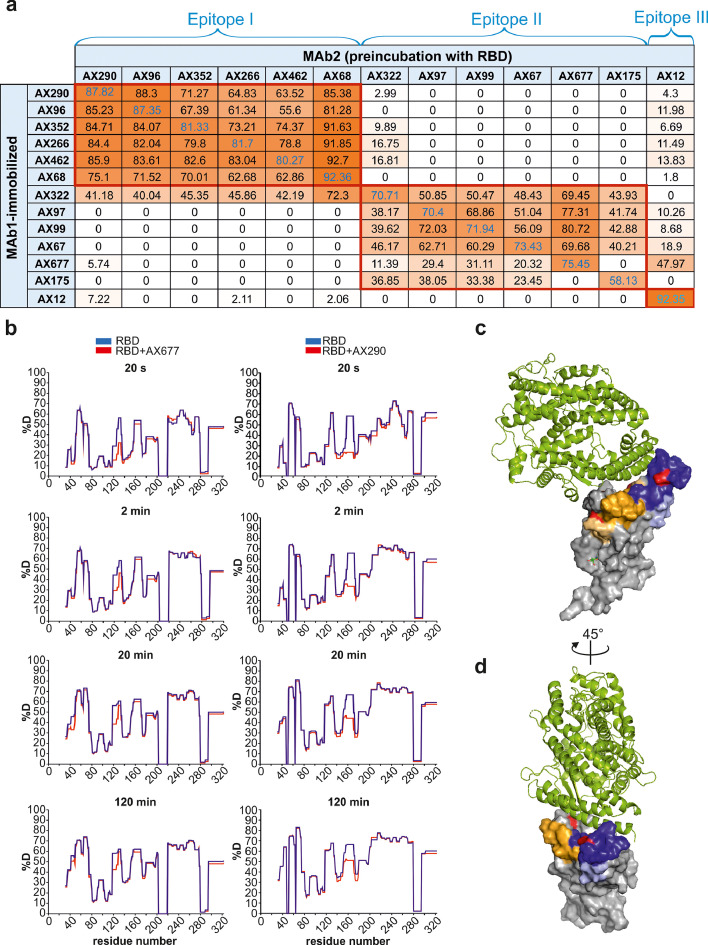
Figure 2.
The panel of selected neutralization antibodies target three non-overlapping epitopes on RBD.
(a) Competitive ELISA was used for determination of the epitopes on RBD by NAbs. Signal reduction for more than 30 % was considered a positive competition. The experiments were performed in duplicates.
(b) Difference plots depicting the changes in deuterium uptake in the RBD peptides upon binding to AX677 (left panels) and AX290 (right panels). The HDX reaction times are indicated in seconds. Numbers on x-axis represent amino acid positions of the recombinant RBD protein (adding 316 aligns them with the S protein numbering). Each plot represents an average of three technical replicates.
(c, d) The positions of the peptides bound by the antibodies and identified in the HDX experiments are highlighted in the structure of SARS-CoV-2 spike receptor-binding domain bound to ACE2 reproduced from PDB 6M0J[41] . The AX290 binding site is represented by shades of blue, AX677 by orange and yellow. Darker colours represent peptides that exhibit reduction in deuterium exchange (%D) ≥20%, light colours represent peptides with reduction in %D between 5-20%. ACE2 is shown in a green cartoon model, RBD as a grey surface model, mutations N439K (within AX677 binding site) and E484K (within AX290 binding site) are shown in red. The model was rendered using PyMOL Molecular Graphics System, Version 2.5.0a0 Open-Source, Schrödinger, LLC.