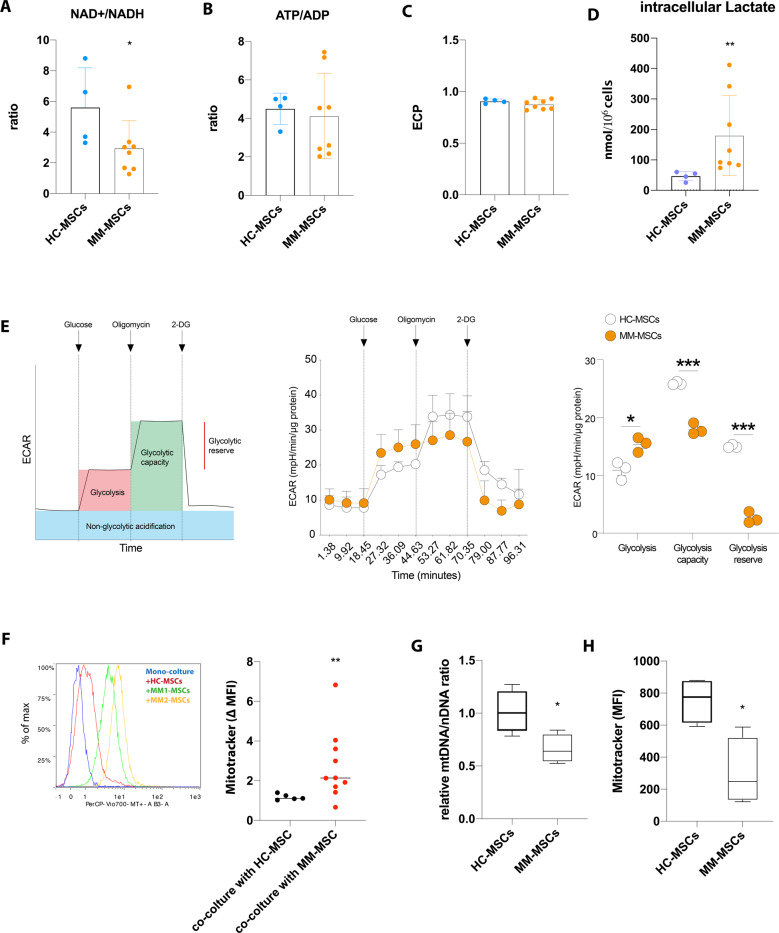
Fig. 1. MM-MSCs are less dependent on mitochondrial metabolism and transfer more mitochondria to myeloma PCs compared to HC-MSCs.
A–C NAD+, NADH, ATP and ADP concentrations were calculated by HPLC analysis in deproteinized HC-MSCs (n = 4) and MM-MSCs (n = 8) and ratio were graphed. ECP was calculated as follows: ATP+1/2 ADP/ATP+ADP+AMP. D Intracellular lactate concentration was measured by enzymatic assay E Representative extracellular acidification rate (ECAR) plot during glycolytic stress test showing non-glycolytic acidification, glycolysis, glycolysis capacity and glycolytic reserve and representative plots of HD-MSCs (n = 3) and MM-MSCs (n = 3) upon glucose, oligomycin, and 2-DG exposition over time. F MitoTracker Red CMXRos uptake by U266 cells (CD45+ gated) was measured by flow cytometry after 24 h co-culture with MitoTracker labeled HC-MSCs (n = 5) or MM-MSCs (n = 10). Δ MitoTracker-mean fluorescence intensity (calculated as the difference between red autofluorescence of U266 cells grown alone and MitoTracker fluorescence of the same cells co-cultured with MSCs) was graphed. A representative flow cytometry histogram shows the comparison of MitoTracker red fluorescence of U266 cells in monoculture vs cells in coculture with HC- or MM-MSCs. G, H Box plots showing mitochondrial content of HC-MSCs (n = 4) and MM-MSCs (n = 4). F Relative quantification of mtDNA levels was performed using qPCR; G mitochondrial mass was evaluated by flow cytometry after MitoTracker staining. Bars indicate the standard error means (*p < 0.05; **p < 0.01; ***p < 0.001).