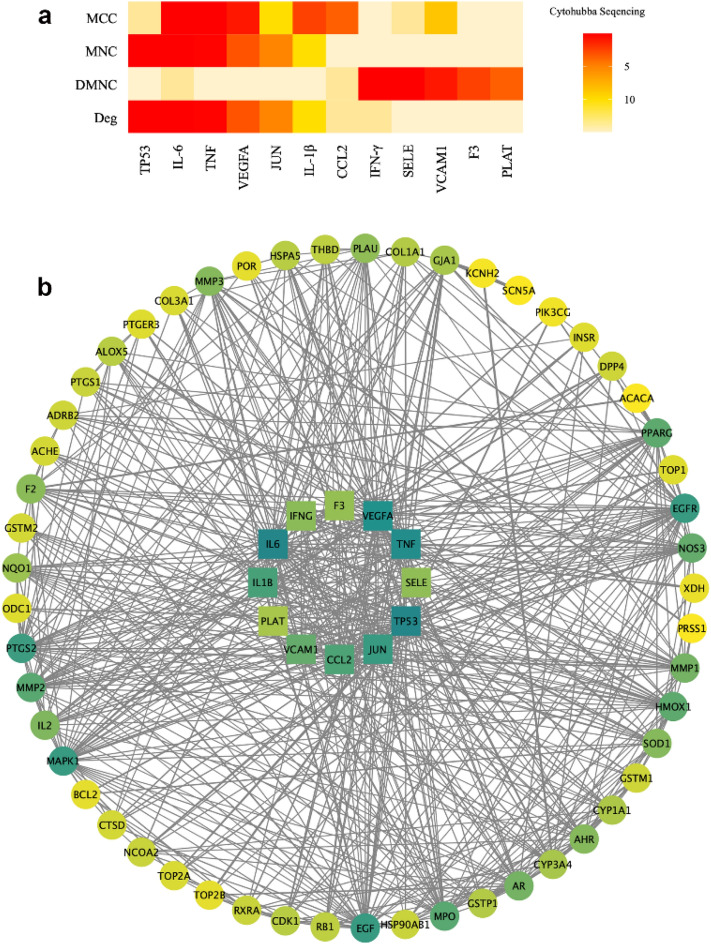
Figure 3.
(a) Topological analysis was performed by the cytohubba tool. The four algorithms, maximum neighborhood component (MNC), density of maximum neighborhood component (DMNC), maximal clique centrality (MCC) and degree (Deg), were used as reference standards. The darker the color, the higher the sequence. The sequence of each common gene in the four algorithms were integrated. The abscissa represented 12 key common targets. (b) This was a protein–protein interaction (PPI) network graph, which showed the interrelationships between the 66 common targets. Each circle represented one common target. The color in the graph was adjusted according to the degree value. The deeper the color, the larger the degree value. The genes in the center of the circle were the 12 key genes that had been calculated using topological analysis.