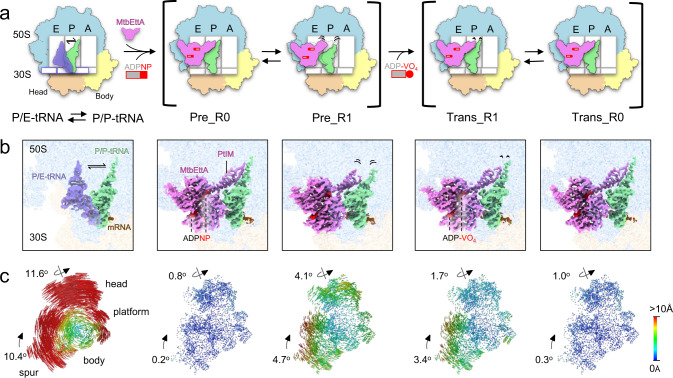
Fig. 1. Cryo-EM structures of Mtb 70SIC in complex with MtbEttA in the pre-hydrolysis and transition states.
a Cartoon representations of Mtb ribosome in different states, labeled with the E, P, and A sites, as well as the 50S and domains (head and body) of the 30S. The first column shows the transition of tRNA between the P/P (light green) and P/E (transparent medium blue) positions. The second and third columns show that MtbEttA (violet), in the Pre_R0 and Pre_R1 states, respectively, binds at the E-site of Mtb 70SIC and interacts with the P-site tRNA. The fourth and fifth columns show the corresponding 70SIC-MtbEttA complexes in the Trans_R1 and Trans_R0 states, respectively. Dashed outlines at the E and P sites (third column) show the position of MtbEttA and the tRNA in the Pre_R0 state. The length of the black curved lines indicates the flexibility in these regions. b Cryo-EM maps showing the density of tRNAs and MtbEttA with the 50S (light blue) and the 30S (light yellow) transparent in the background. PtIM and ATP analogs are labeled. The color scheme follows panel (a). c Analysis of 30S rotations in the P/E-tRNA or MtbEttA-bound states, relative to the non-rotated P/P-tRNA state. The head, body, platform, and spur of the 30S subunit are labeled. Rainbow-colored vectors represent the displacements between equivalent C4’ or Cα atoms of the 30S subunits, with red and blue indicating large and small movements, respectively.