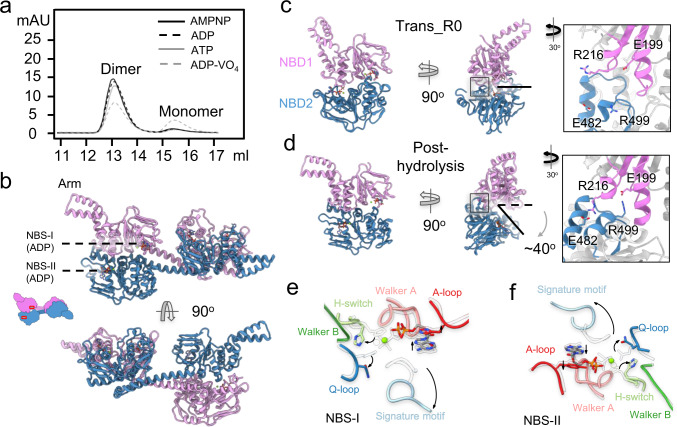
Fig. 6. The crystal structure of MtbEttA in the ADP state.
a Size exclusion chromatography graph indicates the population distribution of dimer and monomer of MtbEttA with different nucleotides. b Crystal structure of MtbEttA bound with ADP. The protomer A (plum) and protomer B (steel blue) form a domain-swapped dimer. ADP molecules are resolved and labeled in both NBSs for each half of the domain-swapped dimer. Conformational differences between the two NBS-forming NBDs in the Trans_R0 (c) and post-hydrolysis (d) states. For the Trans_R0 state, the two NBDs are from the cryo-EM structure of the 70SIC-MtbEttA complex. For the post-hydrolysis state, NBD1 (residues 2-241) and NBD2 (306-550) are from Protomers A and B in the crystal structure, respectively, and form the two NBSs. The direction and degree of NBD2 opening relative to NBD1 are indicated in (d). Insets in c and d show zoom-in views of the interaction between two NBDs. e, f Movements of the conserved motifs in both NBSs from the Trans_R0 state (transparent) to the post-hydrolysis state (colored) are indicated by black curved arrows.