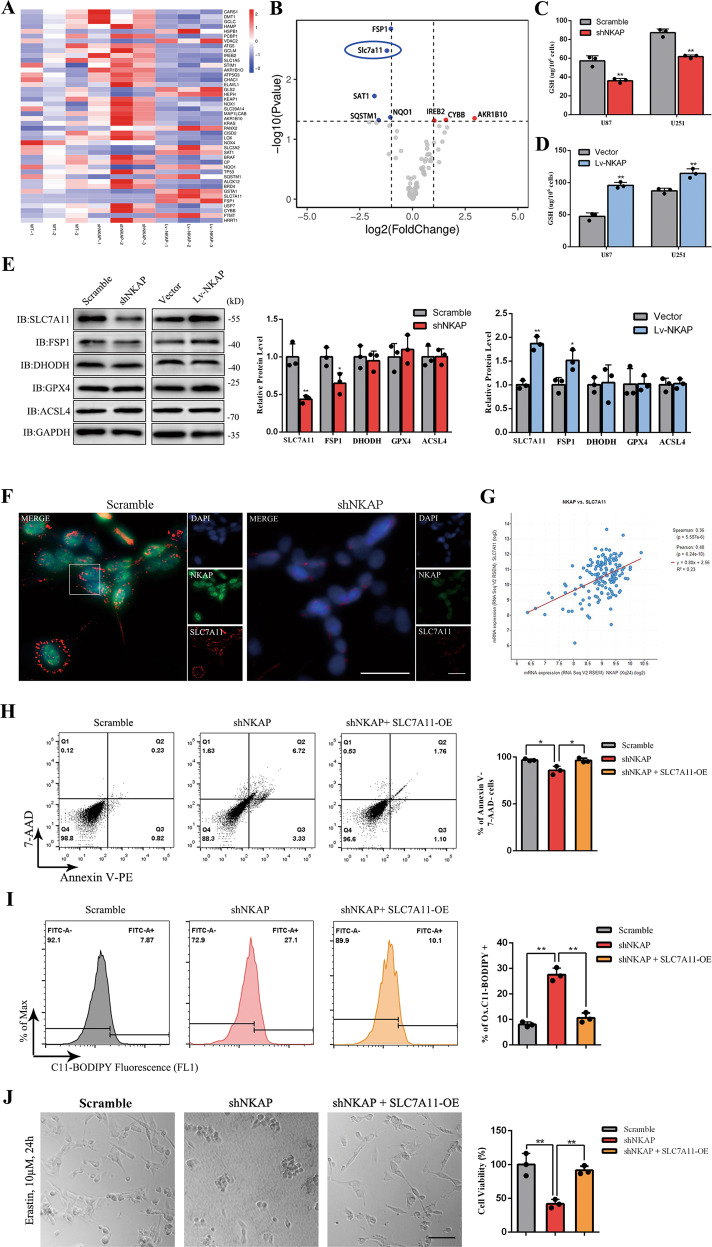
Fig. 3. NKAP controlled ferroptosis by regulating the expression of SLC7A11.
A Ferroptosis-related qPCR array of WT, shNKAP, and Lv-NKAP U87MG cells identified 44 differentially expressed genes. B The volcano map showed the difference in degree and p value, which identified SLC7A11 as the most relevant genes. C GSH level influenced by NKAP knockdown. D GSH level influenced by NKAP overexpression. E Protein levels of ferroptosis core genes measured by western blot assay. GAPDH was used as a loading control. F Immunofluorescence staining showed cell localization and visible expression changes between shNKAP and scramble groups. Scale bar = 50 μm. G Correlation between the expression of NKAP and SLC7A11 in clinical glioblastoma samples was assessed by transcriptome profiling in the TCGA clinical glioblastoma database. H Cell death induced by NKAP knockdown could be blocked by SLC7A11 overexpression detected by flow cytometry in U87MG. I High Lipid peroxidation level induced by NKAP knockdown could be reversed by SLC7A11 overexpression detected by flow cytometry in U87MG. J High drug sensitivity of NKAP knockdown U87MG cells to 10 μM erastin for 24 h could be reversed by SLC7A11 overexpression. Scale bar = 100 μm. All bars show mean ± SD of three independent experiments. *P < 0.05, **P < 0.01.