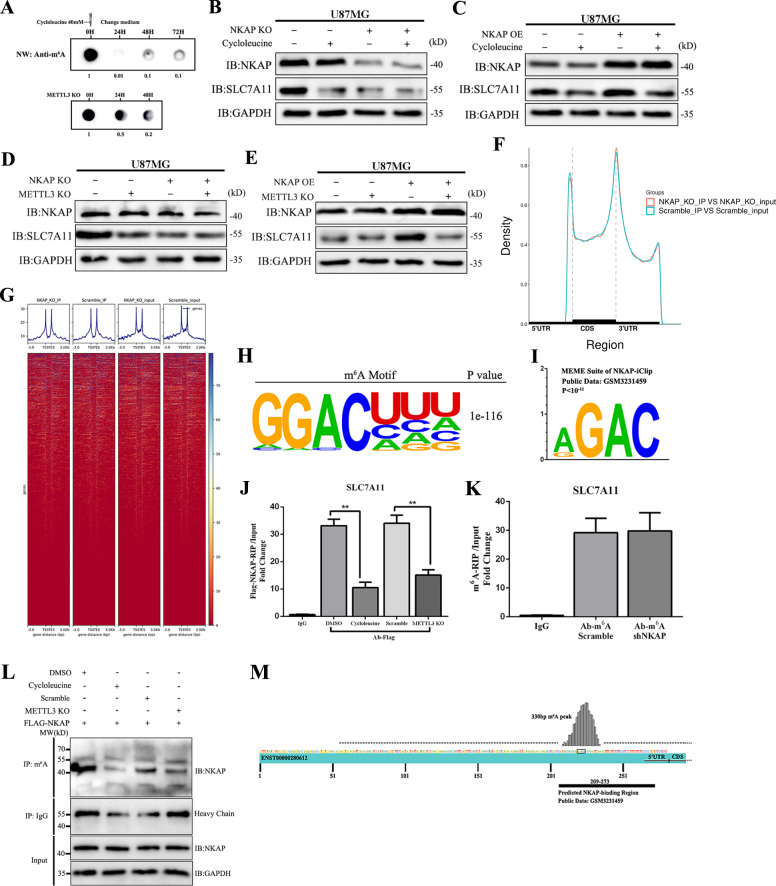
Fig. 5. Binding to m6A was necessary for NKAP to promote SLC7A11 expression.
A m6A northwestern blot assay was performed using an equal amount of RNA (1 μg) isolated from each group. Briefly, the cells were treated with 40 μM cycloleucine for 24 h until the change of culture media. B Western blot assay showed SLC7A11 protein expression regulated by NKAP knockdown and 40 μM cycloleucine for 24 h. C SLC7A11 protein expression regulated by NKAP overexpression and 40 μM cycloleucine for 24 h. D SLC7A11 protein expression is regulated by NKAP knockdown and METTL3 knockdown. E SLC7A11 protein expression regulated by NKAP overexpression and METTL3 knockdown. F Similar m6A peak density in the 5ʹ-UTR, CDS, and 3ʹ-UTR regions in the scramble and shNKAP groups. G Enrichment of reads near the transcription start site (TSS) and transcription end site (TES). H HOMER identified the m6A consensus motif in the U87MG cells as “RGACH”. I MEME SUIT analyzed NKAP footprints as the “RGAC” motif based on the NKAP-iCLIP public data. J FLAG-NKAP-RIP revealed that enrichment of SLC7A11 mRNA significantly decreased in the cycloleucine-treated and METTL3 knockdown groups. K m6A-RIP showed no significant difference in SLC7A11 mRNA enrichment between the scramble and shNKAP groups. L Co-IP analysis of the interaction between m6A and Flag-NKAP in U87MG cells. M m6A-seq showed that the m6A peak in the SLC7A11 transcript ENST00000613322 had an overlap with the predicted NKAP binding region. GAPDH was used as an internal control in western blot. All bars show mean ± SD of three independent experiments. *P < 0.05, **P < 0.01.