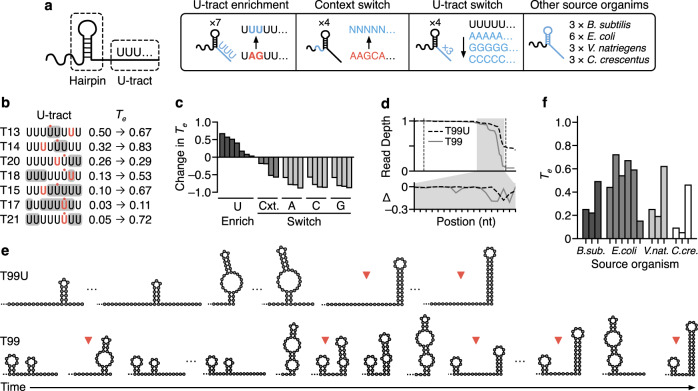
Fig. 5. Exploring design features of the core terminator.
a Overview of modifications made to core terminators, tested without spacers or modifiers: (left to right) U-tract enrichment to include up to 8 consecutive U residues down-stream of the core terminator hairpin; Context switch with the 19–25 nt immediately upstream of the terminator hairpin changed to a random sequence; U-tract switch to consecutive A, C, or G residues; Other source organisms used to find diverse terminator sequences. b Effect of U-tract enrichment on most common termination position and change in termination efficiency (Te). Substitution of nucleotides for U are indicated with gray shading. Termination position before the substitution is indicated in bold red text. Termination position after substitution is indicated with red dot. c Change in Te for U-tract enrichment (U Enrich, dark gray), context switch (Cxt, gray) and U-tract switches (A, C, G, light gray). d Normalized dRNA-seq read depth profiles for the T7 phage T-theta terminator (T99, gray solid line) and a variant (T99U, black dashed line). Dotted lines denote the start and end of the core terminator. The gray-shaded region is expanded in the lower panel showing the change in normalized read depth at each nucleotide position with respect to the previous nucleotide (Δ). e Secondary structures predicted by co-transcriptional folding simulation of terminator formation (left to right) for T99U and T99. Positions where measured termination occurs are indicated with red triangles. f Measured Te of T7 RNAP for terminators sourced from diverse organisms. Source data are provided as a Source Data file.