Abstract
Background
There are few clinically useful circulating biomarkers of lung function and lung disease. We hypothesized that genome-wide association studies (GWAS) of circulating proteins in conjunction with GWAS of pulmonary traits represents a clinically relevant approach to identifying causal proteins and therapeutically useful insights into mechanisms related to lung function and disease.
Study Question
Can an integrative genomic strategy using GWAS of plasma soluble receptor for advanced glycation end-products (sRAGE) levels in conjunction with GWAS of lung function traits identify putatively causal relations of sRAGE to lung function?
Study Design and Methods
Plasma sRAGE levels were measured in 6,861 Framingham Heart Study participants and GWAS of sRAGE was conducted to identify protein quantitative trait loci (pQTL), including cis-pQTL variants at the sRAGE protein-coding gene locus (AGER). We integrated sRAGE pQTL variants with variants from GWAS of lung traits. Colocalization of sRAGE pQTL variants with lung trait GWAS variants was conducted, and Mendelian randomization was performed using sRAGE cis-pQTL variants to infer causality of sRAGE for pulmonary traits. Cross-sectional and longitudinal protein-trait association analyses were conducted for sRAGE in relation to lung traits.
Results
Colocalization identified shared genetic signals for sRAGE with lung traits. Mendelian randomization analyses suggested protective causal relations of sRAGE to several pulmonary traits. Protein-trait association analyses demonstrated higher sRAGE levels to be cross-sectionally and longitudinally associated with preserved lung function.
Interpretation
sRAGE is produced by type I alveolar cells, and it acts as a decoy receptor to block the inflammatory cascade. Our integrative genomics approach provides evidence for sRAGE as a causal and protective biomarker of lung function, and the pattern of associations is suggestive of a protective role of sRAGE against restrictive lung physiology. We speculate that targeting the AGER/sRAGE axis may be therapeutically beneficial for the treatment and prevention of inflammation-related lung disease.
Key Words: COPD, Mendelian randomization, spirometry, sRAGE
Abbreviations: AGER, advanced glycation end-products receptor gene; FHS, Framingham Heart Study; GWAS, genome-wide association study; MMP, matrix metalloproteinase; MR, Mendelian randomization; %LAA-950, percentage of low attenuation areas below −950 HU as measured by CT; PP, posterior probability; pQTL, protein quantitative trait locus; SNP, single-nucleotide polymorphism; sRAGE, soluble receptor for advanced glycation end-products
FOR EDITORIAL COMMENT, SEE PAGE 3
Identifying causal variants, genes, proteins, and biological pathways for complex diseases is a major challenge of translational research,1 because genome-wide association studies (GWAS) of these phenotypes often yield variants that have little or no apparent biological relevance.2 In contrast, GWAS of circulating levels of proteins, as gene products, are more likely to identify genetic associations that are mechanistic in nature and can serve as clinically relevant drug targets.3 As part of the Systems Approach to Biomarker Research in Cardiovascular Disease,4 we assayed and conducted a GWAS of 71 plasma proteins in 6,861 Framingham Heart Study (FHS) participants and demonstrated that an integrative genomics approach can identify putatively causal protein biomarkers for cardiovascular disease.5 We postulate that this approach can be extended to other complex diseases. For proof-of-concept, we explored the soluble receptor for advanced glycation end-products (sRAGE) as a causally protective protein in relation to lung function.
COPD and its anatomic and radiographic correlate, pulmonary emphysema, constitute the third leading cause of nonaccidental death in the United States.6 COPD is diagnosed using spirometry,7 and emphysema is identified by lung imaging, including CT, which measures the percentage of emphysema-like lung.8 COPD has environmental risk factors such as smoking9 and genetic risk factors such as polymorphisms within SERPINA1 that increase the risk of an inherited form of lung disease due to α1-antitrypsin deficiency.8 Here, we focus on the analyses of plasma sRAGE in relation to lung function by spirometry and lung structure by CT imaging. sRAGE has previously been explored as a biomarker of COPD and emphysema.10 Genetic variants in the AGER gene, which encodes RAGE, have been reported to be associated with circulating sRAGE levels and with impaired lung function, COPD, and emphysema,11,12 but a causal link in humans between sRAGE and lung function or lung disease has not been proven.
To explore a causal relation of sRAGE to lung function and structure, we carried out a comprehensive integrative genomic study of genetic variants (pQTLs; protein quantitative trait loci) associated with plasma levels of sRAGE5 in conjunction with GWAS of lung traits13,14 and tested for colocalization of these GWAS signals and performed Mendelian randomization (MR) analyses to infer causality of sRAGE in relation to pulmonary traits. Further evidence implicating sRAGE is provided from the results of cross-sectional and longitudinal analyses of sRAGE in relation to pulmonary traits in FHS participants. Our findings, in conjunction with prior in vivo and in vitro functional studies, provide an in-depth exploration of the hypothesis that the advanced glycation end-products receptor gene (AGER)/sRAGE axis is causally related to lung function and structure and that sRAGE is causally protective.
Methods
Study Design
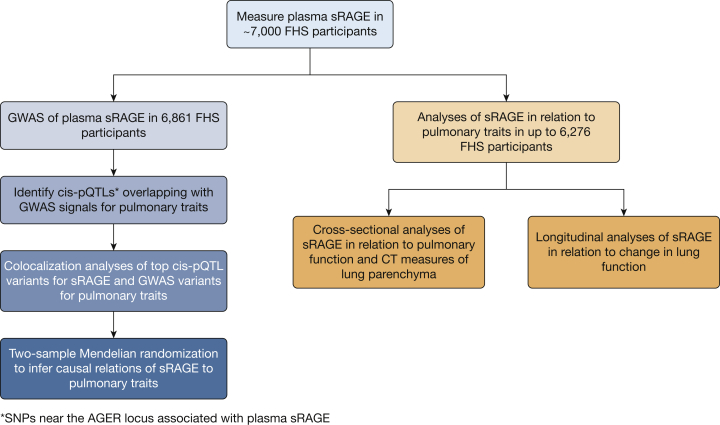
The study consisted of five steps (Fig 1). First, our GWAS results for sRAGE (e-Table 1), performed on 6,861 FHS participants with measured plasma sRAGE levels and genome-wide single-nucleotide polymorphism (SNP) genotyping,5 using Affymetrix 500K mapping arrays and imputed to the 1000 Genomes Project reference panel (build 37 phase 1 v3),15 were interrogated to identify pQTL variants associated with circulating sRAGE levels. Second, all cis-pQTL variants for sRAGE (e-Table 1), defined as within 1 megabase (Mb) of the transcription start site of the RAGE protein coding gene, AGER,5 were interrogated to identify those that overlap with SNPs from previously published GWAS of lung traits.14,15 Third, colocalization analyses were performed to infer shared genetic signals between sRAGE and lung traits. Fourth, using sRAGE cis-pQTL variants as instrumental variables, two-sample MR5 was conducted to infer causality of sRAGE in relation to lung traits. Fifth, protein-trait association analyses were performed in FHS participants using plasma sRAGE measurements and cross-sectional and longitudinal spirometry measures of pulmonary function and CT scan measures of pulmonary parenchymal density.
Figure 1.
This study consisted of multiple steps: measurement of plasma sRAGE in ~7,000 FHS participants; identification of single-nucleotide polymorphisms (SNPs) associated with circulating sRAGE (ie, pQTLs; protein quantitative trait loci); identification of sRAGE cis-pQTL variants that overlapped with previously published COPD/emphysema GWAS SNPs; colocalization analysis to infer shared genetic variants for sRAGE and COPD/emphysema; Mendelian randomization to infer putative causality of sRAGE for emphysema; cross-sectional and longitudinal analyses of sRAGE in relation to pulmonary traits in FHS participants.
Colocalization
We sought to determine whether pQTL variants associated with circulating sRAGE levels colocalized with SNPs associated with relevant pulmonary traits, including FEV1, FVC, the ratio FEV1/FVC, and the percentage of emphysematous-like lung parenchyma on CT imaging (percentage of low attenuation areas at −950 Hounsfield units [%LAA-950]) from prior GWAS.14,16 To do this, we first identified all cis-pQTL variants for plasma sRAGE levels. Using GWAS SNPs associated with pulmonary traits from the entire sRAGE cis-pQTL locus (1 Mb upstream and downstream),13 we conducted a Bayesian test for colocalization of sRAGE cis-pQTL variants and pulmonary function GWAS SNPs using the COLOC version 3.1 package in R.17 Briefly, this method calculates Bayes factors for five different hypotheses—no SNPs associated with sRAGE or pulmonary function (H0), SNPs associated with sRAGE only (H1), SNPs associated with pulmonary function only (H2), two distinct SNPs with one for sRAGE only and the other for pulmonary function only (H3), and one distinct SNP for sRAGE and pulmonary function (H4)—within a genomic region by integrating all binary SNP configurations (ie, each SNP is responsible or not responsible for the observed association signal) that support each hypothesis. Prior probabilities were specified for an SNP being associated with plasma sRAGE levels only (p1), pulmonary trait only (p2), and pulmonary trait given its association with sRAGE (p12). p1 and p2 were set to .0001, and p12 was then set to .00001, estimating that 1 in 10,000 SNPs were associated with each trait. Posterior probabilities (PP) were then calculated for each of the five hypotheses, with PPH4 indicating the posterior probability of colocalization (one distinct variant associated with sRAGE and the pulmonary trait).
Mendelian Randomization
sRAGE cis-pQTL variants were pruned at a linkage disequilibrium threshold of r2 < 0.1, estimated from the European samples within the 1000 Genomes Project using PLINK.18 The resulting pruned cis-pQTL variants, along with their associations with plasma sRAGE levels (in FHS), were uploaded to MR-base19 as the exposure, and pulmonary traits from prior GWAS (FEV1, FVC, FEV1/FVC, prevalent COPD, and %LAA-950) were selected as the outcomes.16,20 MR inverse-variance weighted and Egger regression were conducted twice—the first time using all nonredundant sRAGE cis-pQTL variants (linkage disequilibrium r2 < 0.1; N = 9) as instrumental variables and repeated using the same set of nonredundant sRAGE cis-pQTL variants excluding rs2070600 (Gly82Ser), a nonsynonymous variant at the ligand binding domain, with the Ser82 allele having nearly a twofold higher binding affinity (KD for Ser82 = 77 nM; KD for Gly82 = 122 nM).21 MR causal effect estimates were reported per standard error increment in inverse-rank normalized plasma sRAGE.
Study Population
The study sample for protein-trait association analysis (n = 6,276) consisted of FHS offspring (n = 2,512) and third generation (n = 3,764) cohort participants with plasma sRAGE measurements and pulmonary function measurements at the baseline examination (offspring cohort examination 7 [1999-2003] and third generation cohort examination 1 [2002-2005]; Table 1). For the longitudinal analyses, participants who did not attend the follow-up examination (offspring cohort examination 8 and third generation cohort examination 2) or had COPD, defined as FEV1/FVC < 0.7 at the baseline examination22 (n = 1,161), were excluded. The final longitudinal study sample consisted of 4,136 FHS participants (e-Table 2). Secondary protein-spirometry trait analyses were conducted on FHS participants who were stratified by baseline smoking status (current, former, and never smokers; e-Table 3). Protein-trait association analyses were also conducted on 2,707 participants who had multi-detector CT scan measurement of emphysematous lung parenchyma (%LAA-950; e-Table 4).
Table 1.
Baseline Clinical Characteristics of FHS Participants With Spirometry Measures of Pulmonary Function (N = 6,276)
| Characteristic | Mean | SD |
|---|---|---|
| Age, y | 48.3 | 13.4 |
| Height, in | 66.7 | 3.72 |
| BMI | 27.4 | 5.42 |
| Pack-years of smoking | 9.27 | 16.71 |
| Plasma sRAGE, pg/mL | 3,585 | 1,150 |
| FEV1, L | 3.22 | 0.90 |
| FVC, L | 4.24 | 1.09 |
| FEV1/FVC | 0.76 | 0.074 |
| No. | % | |
| Women | 3,360 | 53.5 |
| COPD | 1,161 | 18.5 |
| Current smoker | 979 | 15.6 |
| Never smoker | 3,266 | 52.0 |
| Former smoker | 2,031 | 32.4 |
sRAGE = soluble receptor for advanced glycation end-products.
Clinical Measures
Continuous spirometry measures of pulmonary function included FEV1, FVC, and FEV1/FVC obtained at the same examination at which circulating sRAGE was measured. Chest CT was used to assess lung parenchymal density.23 The demographic and clinical characteristics of the CT sample are shown in e-Table 4. Lung parenchymal density was analyzed as a continuous variable, defined as the percentage of low attenuation areas (below −950 Hounsfield units) as measured by CT (%LAA-950)24; a high value is indicative of greater emphysematous lung parenchyma.
Protein Quantification
Baseline plasma sRAGE concentration was measured as part of the Systems Approach to Biomarker Research in Cardiovascular Disease Initiative. Briefly, fasting blood plasma samples were collected from FHS participants at the baseline examination (offspring cohort examination 7 [1999-2003] and third generation cohort examination 1 [2002-2005]; Table 1) and stored at −80 °C. Plasma sRAGE was quantified using a modified enzyme-linked immunosorbent assay, multiplexed on a Luminex xMAP platform (Sigma-Aldrich), using previously published methods.25 To avoid cross-reactivity, all targets were initially developed as singleton assays before compatible targets were combined to form multiplex panels. For the sRAGE assay, the detection antibody was MAB11451 (R&D Systems), the capture antibody was product 1145-RG-050 (R&D Systems), and the reference protein was BAF1145 (R&D Systems). The mean inter- and intra-assay coefficients of variation for sRAGE were 5.6% and 14.5%, respectively.4
Statistical Methods
Statistical analyses were performed using R software version 3.1.126 and SAS software version 9.4. For all protein-trait association analyses, plasma sRAGE levels were log-transformed and standardized around a mean of 0 and SDof 1. For the cross-sectional analyses of pulmonary function, baseline plasma sRAGE levels were analyzed in relation to FEV1, FVC, FEV1/FVC, and COPD, after adjusting for age, sex, height, pack-years of smoking, study cohort, and current and former smoking status at the baseline examination. For the cross-sectional analyses of sRAGE in relation to CT measures of lung function, baseline plasma levels were analyzed in relation to %LAA-950 after adjusting for age, sex, BMI, study cohort, current and former smoking status, and smoking pack-years at the examination closest to the time of the CT scans. For the longitudinal analyses of changes in pulmonary function, baseline plasma sRAGE levels were analyzed in relation to follow-up minus baseline spirometry values: ΔFEV1, ΔFVC, Δ(FEV1/FVC, and incident COPD, after adjusting for baseline age, sex, height, pack-years of smoking, study cohort, smoking status, and baseline pulmonary function. A positive regression coefficient in the longitudinal protein-trait analysis was interpreted as higher baseline sRAGE being associated with a longitudinal increase in the respective spirometry measure (eg, a protective effect). Generalized estimating equations were applied to account for familial correlations among FHS participants. Secondary protein-trait analyses were stratified by smoking status.
Study Approval
Informed consent for genetic research was given by all participants in this investigation. The study protocol was approved by the Boston University Medical Center Institutional Review Board (protocol H-27984).
Results
Baseline Characteristics
The demographic and clinical characteristics of the FHS study participants in the cross-sectional spirometry analyses are shown in Table 1. The total sample for the cross-sectional analyses of sRAGE in relation to spirometry measures of pulmonary function included 6,276 FHS participants (mean age, 48 years; 54% women); 2,512 from the offspring and 3,764 from the third generation cohort. For the longitudinal spirometry analyses, participants who did not attend the follow-up examination and those with COPD at baseline were excluded, leaving a final study sample of 4,136 (mean baseline age, 46 years, 55% women; e-Table 2). Baseline clinical characteristics of the cross-sectional study sample, stratified by smoking status, are provided in e-Table 3. Characteristics of the study sample with CT assessment of emphysematous lung parenchyma (%LAA-950; n = 2,707) are provided in e-Table 4.
Colocalization Analyses
Colocalization of genetic signals for circulating sRAGE and lung traits at rs2070600 within the AGER locus was significant for FVC (PPH4 = 99.8%), FEV1/FVC (PPH4 = 99.9%), and %LAA950 (PPH4 = 99.9%), but not for FEV1 (PPH4 = 2.56E-16; e-Table 5).
MR Causal Inference Testing
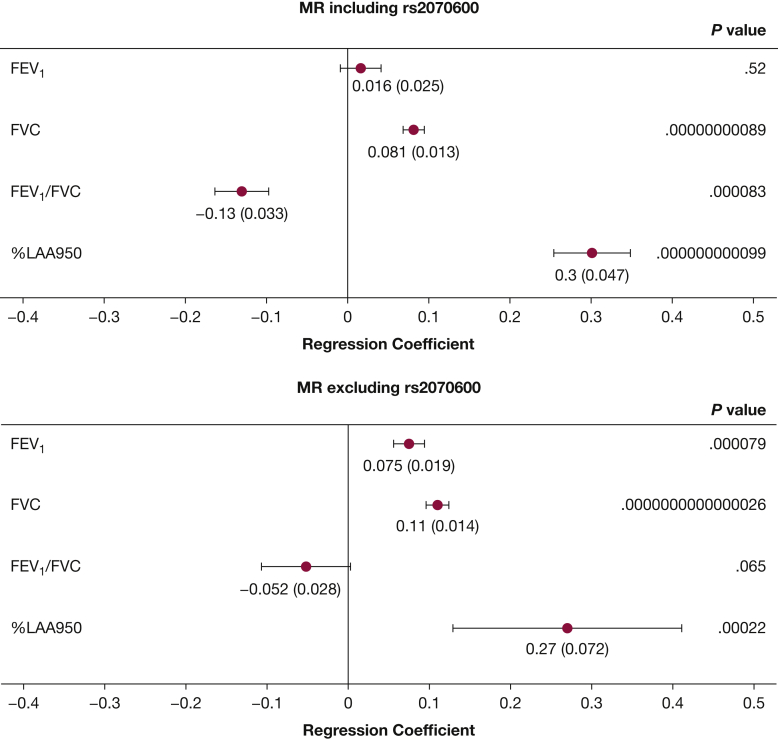
MR results for causal inference of the relationship between sRAGE and pulmonary traits are summarized in Figure 2. Primary MR analyses using all nonredundant sRAGE cis-pQTL variants (n = 9 SNPs) demonstrated sRAGE to be causal for FVC (β = 0.081, P = .00000000089), FEV1/FVC (β = −0.13, P = .000083), and %LAA950 (β = 0.30, P = .000000000099; Fig 2, top). MR analyses excluding missense variant rs2070600 (n = 8 SNPs) demonstrated similar results for FVC and %LAA950, whereas FEV1/FVC was no longer statistically significant (Fig 2, bottom). MR analyses for COPD were not statistically significant whether including (OR = 1.08, P = .31) or excluding (OR = 0.91, P = .055) rs2070600. Forest plots showing individual effects for each SNP used as an instrumental variable in MR analyses for FEV1 and FVC (e-Fig 1) demonstrated that rs2070600 significantly drove the MR association of sRAGE with FEV1. MR Egger regression analyses (e-Table 6) including rs2070600 demonstrated statistically significant Egger intercepts for FEV1 (P = .000833), FVC (P = .00397), and FEV1/FVC (P = .00949), whereas analyses excluding rs2070600 resulted in nonsignificant Egger intercepts, further suggesting that rs2070600 significantly drove the MR results.
Figure 2.
Results from MR analyses conducted including rs2070600 (top) and excluding rs2070600 (bottom). SNPs (apart from rs2070600) used as IVs included: rs11796, rs1871665, rs204993, rs2073044, rs2170185, rs2523570, rs6923504, and rs9266529. SNP = single-nucleotide polymorphism. ∗SNP-outcome associations are from GWAS from Shrine et al.16 Regression coefficients for FEV1 and FVC are reported as change in inverse rank-normalized lung volumes per SD increase in inverse rank-normalized plasma sRAGE. Regression coefficients for FEV1/FVC are reported as change in FEV1/FVC per SD increase in inverse rank-normalized plasma sRAGE. ∗∗SNP-outcome associations are from Cho et al14 %LAA-950 was defined as the percentage low attenuation areas (below −950 Hounsfield units) by chest CT. Regression coefficients for %LAA-950 are reported as change in log-transformed %LAA-950 per SD increase in inverse rank-normalized plasma sRAGE. Regression coefficients for FEV1 and FVC are reported as change in inverse rank-normalized lung volumes per SD increase in inverse rank-normalized plasma sRAGE. Regression coefficients for FEV1/FVC are reported as change in FEV1/FVC per SD increase in inverse rank-normalized plasma sRAGE. Regression coefficients and SD are shown below each corresponding forest plot point estimate, formatted as beta (SD). All statistically significant P values (right column) are shown in bold.
Protein-Trait Association Analysis; Cross-Sectional and Longitudinal Associations of sRAGE With Pulmonary Traits
Plasma sRAGE was associated cross-sectionally with FEV1 (β = 0.021, P = .00054), FVC (β = 0.038, P = .000000061), and FEV1/FVC (β = −0.21, P = .017), but not prevalent COPD (OR = 0.95, P = .20). Results of the cross-sectional association analyses between sRAGE and pulmonary traits are shown in Table 2. Cross-sectional association between sRAGE and CT measures of lung parenchymal density showed higher sRAGE to be associated with lower %LAA-950 (β = −0.0017, P = .0012), consistent with a protective effect of sRAGE on the lung parenchyma (Table 2). Longitudinal analyses revealed that baseline plasma sRAGE was associated with greater ΔFEV1 (β = 2.26, P = .00025), greater ΔFVC (β = 4.2, P = .0000000090), but lower Δ(FEV1/FVC) (β = −0.026, P = .0018, Table 3). Cross-sectional protein-trait association analyses between sRAGE and pulmonary traits, when stratified by smoking status, remained directionally consistent with the primary analyses, but sRAGE had a larger (protective) effect size in current smokers than in former and never smokers on FEV1 (β = 0.059, 0.031, and 0.036, respectively) and FVC (β = 0.087, 0.054, and 0.064; e-Table 7).
Table 2.
Observed Cross-sectional Associations Between sRAGE and Pulmonary Traits
| Cross-sectional Trait | Cross-sectional Association With sRAGE |
||
|---|---|---|---|
| β/OR | SE/95% CI | P | |
| FEV1a | 0.021 | 0.006 | .000537 |
| FVCa | 0.038 | 0.007 | .0000000614 |
| FEV1/FVCb | −0.21 | 0.088 | .0165 |
| Prevalent COPD | OR, 0.95 | 95% CI, 0.88-1.02 | .195 |
| %LAA-950b | −0.002 | 0.000 | .000101 |
Covariates for FEV1, FVC, FEV1/FVC, and COPD: age, sex, height (inches), pack-year of smoking, study cohort, current smoker (yes/no), former smoker (yes/no). COPD was defined as FEV1/FVC < 0.7. ORs for COPD are reported per 1 SD increase in log-transformed sRAGE concentration. Statistically significant P values are shown in bold. %LAA-950 = percentage low attenuation area (below −950 Hounsfield units); sRAGE = soluble receptor for advanced glycation endproducts.
FEV1 and FVC are expressed in L.
Analyses for %LAA950 were additionally adjusted for BMI. A negative coefficient for %LAA-950 reflects less emphysema-like lung in relation to higher sRAGE levels.
Table 3.
Observed Longitudinal Associations Between sRAGE and Pulmonary Traits
| Longitudinal Trait | Longitudinal Association With sRAGE |
||
|---|---|---|---|
| β/OR | SE/95% CI | P | |
| ΔFEV1a | 2.256 | 0.616 | .000251 |
| ΔFVCa | 4.168 | 0.725 | .00000000895 |
| Δ(FEV1/FVC)b | −0.026 | 0.008 | .00176 |
| Incident COPDc | OR, 1.02c | 95% CI, 0.88-1.17 | .830 |
Covariates in longitudinal analyses included age, sex, height (inches), pack-year of smoking, study cohort, current smoker (yes/no), former smoker (yes/no), baseline pulmonary function. Longitudinal regression coefficients reflect follow-up minus baseline differences in continuous spirometry values per SD increment in standardized long-transformed sRAGE. A positive regression coefficient for spirometry traits reflects greater preservation of a spirometry measure over time in relation to higher sRAGE levels. Statistically significant P values are shown in bold.
Regression coefficients for ΔFEV1 and ΔFVC are reported as change in lung volume (mL; at follow-up minus baseline) per SD increase in standardized log-transformed plasma sRAGE.
Regression coefficients for FEV1/FVC and %LAA-950 are reported as per SD increase in standardized log-transformed plasma sRAGE.
ORs for COPD are reported per SD increase in standardized log-transformed plasma sRAGE. COPD was defined as FEV1/FVC < 0.7.
Discussion
We provide evidence from an integrative genomics approach that sRAGE is a putatively causal and protective protein biomarker in relation to lung function. MR analyses revealed higher sRAGE levels to be causal and protective in relation to FVC, and protein-trait analyses in our large study sample demonstrated higher sRAGE levels to be associated with higher FVC, consistent with the protective effects seen in MR (Fig 2 and Table 2). Longitudinal analyses similarly revealed that higher baseline sRAGE levels were associated with preservation of FEV1 and FVC over time (Table 3), suggesting a longitudinal protective effect of sRAGE on lung function. Our spirometry results for the association of sRAGE with FEV1 and FVC remained significant in separate analyses of current, former, and never smokers (e-Table 7). The larger effect sizes observed in current smokers, however, suggested that the protective effect of sRAGE on pulmonary function is greatest among current smokers in whom the pro-inflammatory stimulus for lung parenchymal damage is greatest. Indeed, a prior mouse study demonstrated that the protective effects of knocking out AGER were only observed in mice exposed to cigarette smoke.27 Of note, a recent RNA-seq analysis of AGER expression in bronchial biopsies demonstrated an up-regulation in sRAGE generation via alternative splicing in smokers as compared with never smokers.28
Prior studies have shown that RAGE contributes to lung inflammation,29 which is a contributing factor to lung disease, including COPD,30 pulmonary emphysema,31 and pulmonary fibrosis.32 Membrane-associated RAGE (mRAGE) functions as a pro-inflammatory receptor stimulating nuclear factor kappa-B, mitogen-activated protein kinases, and oxidases.33 Conversely, the soluble form of RAGE, sRAGE, has the opposite effect (ie, antiinflammatory), as it acts as a decoy receptor for RAGE ligands,34 which is also supported by our findings. sRAGE is generated by dual processes: matrix metalloproteinase (MMP)-mediated ectodomain shedding of mRAGE and alternative splicing and removal of AGER exon 10, which encodes the transmembrane domain of the RAGE protein,35 resulting in endogenously secreted RAGE (esRAGE).36 Notably, rs1973612, an SNP in the KLKB1 gene locus, is a trans-pQTL both for sRAGE5 and MMP-2,37 providing a potential genetic link between sRAGE, cleavage of membrane-bound RAGE by MMPs, and lung parenchymal damage in emphysema and pulmonary fibrosis.32,38
RAGE is expressed at basal levels in diverse cell types and tissues, but it is highly expressed in lung tissue and particularly in type I pulmonary alveolar cells.33 Addition of exogenous sRAGE was shown to ameliorate pulmonary inflammation in a rat model of lipopolysaccharide-induced inflammation,39 a mouse model of acid-induced lung injury,40 and multiple mouse models of allergic airways disease.41,42 Therefore, factors that raise circulating levels of sRAGE may diminish the pulmonary inflammatory response to cigarette smoke and other pro-inflammatory stimuli. Considered as a whole, our results, in concert with prior animal experiments, indicate that sRAGE is protective against progressive loss of lung function, particularly in response to inflammation, such as that caused by cigarette smoke exposure.
We acknowledge several limitations of our study. First, our study participants were of European ancestry, and therefore, our results may not be generalizable to other ethnicities. Second, our study was not adequately powered to assess the prognostic utility of plasma sRAGE as a biomarker of dichotomous traits such as new-onset COPD or emphysema. Third, although sRAGE was positively associated with both FEV1 and FVC—consistent with preservation of both measures of lung function—the direction of association between sRAGE and FEV1/FVC was negative both in MR and protein-trait association analyses. Based on these findings, we hypothesize that sRAGE exerts a greater protective effect against restrictive than against obstructive lung disease, as reflected in the greater longitudinal preservation of FVC (β = 4.17) than of FEV1 (β = 2.26; Table 3). Indeed, colocalization analyses also suggested shared genetic pathways between sRAGE and FVC (PPH4 = 99.9%, e-Table 5), but not with FEV1 (PPH4 = .0000000000000256, e-Table 5). Of note, a prior study demonstrated that lungs from Ager-/- mice had greater compliance and lower elastin mRNA and protein levels, but no significant changes in airway resistance.43 Finally, analyses of CT emphysema-like lung density (%LAA-950) also did not provide evidence that sRAGE is protective against emphysema; rather, the body of evidence from our study suggests that sRAGE is a causal and protective biomarker in relation to restrictive rather than obstructive lung disease.
To our knowledge, this is the first study to use an integrative genomics strategy to establish sRAGE as a causal and protective protein biomarker of lung function. We have shown that sRAGE is protective against loss of lung function and provided evidence that the AGER/RAGE axis is a promising therapeutic target for the treatment and prevention of lung damage caused by inflammation, such as that caused by cigarette smoking. Overall, we have demonstrated the applicability of our integrative approach to elucidate putatively causal biomarkers for complex disease phenotypes that can be explored for therapeutic and prognostic utility.
Interpretation
sRAGE is produced by type I alveolar cells and acts as a decoy receptor to block the nuclear factor kappa-B inflammatory signaling cascade. Our findings provide mechanistic evidence from an integrative genomics approach that sRAGE has a causal and protective effect on lung function and that targeting the AGER/sRAGE axis may be therapeutically beneficial for the treatment and prevention of inflammation-related lung disease.
Take-home Points.
Study Question: Can an integrative genomic strategy using GWAS of plasma soluble receptor for advanced glycation end-products (sRAGE) levels in conjunction with GWAS of lung function traits identify putatively causal relations of sRAGE to lung function?
Results: Colocalization identified shared genetic signals for sRAGE with lung traits. Mendelian randomization analyses suggested protective causal relations of sRAGE to several pulmonary traits. Protein-trait association analyses demonstrated higher sRAGE levels to be cross-sectionally and longitudinally associated with preserved lung function.
Interpretation: sRAGE is produced by type I alveolar cells, and it acts as a decoy receptor to block the inflammatory cascade. Our integrative genomics approach provides evidence for sRAGE as a causal and protective biomarker of lung function, and the pattern of associations is suggestive of a protective role of sRAGE against restrictive lung physiology. We speculate that targeting the AGER/sRAGE axis may be therapeutically beneficial for the treatment and prevention of inflammation-related lung disease.
Acknowledgement
Author contributions: J. K., C. Y., and D. L. conceived and designed the research; P. C., E. K. S., and D. L. supervised the experiments; J. K., C. Y., and J. P. M. wrote the manuscript; S.-J. H., C. Y., and J. K. analyzed the data. S.-J. H., J. D., G. C., G. W., and M. H. C. contributed statistical advice, or analysis tools; all authors discussed the results and reviewed the final manuscript.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: E. K. S. received institutional grant support from GSK and Bayer. The National Heart, Lung, and Blood Institute and Ionis Pharmaceuticals entered into a Cooperative Research and Development Agreement (CRADA) to conduct research targeting the AGER gene in relation to lung disease, based in part on the results of this research. D. L. is the NHLBI principal investigator on the CRADA. Neither D. L. nor the NHLBI is receiving any funding from Ionis Pharmaceuticals in conjunction with the CRADA. None declared (J. K., C. Y., S.-J. H., P. C., G. Y. L., J. D., J. P. M., G. O., G. R. W., M. H. C.)
Other contributions: The authors acknowledge manuscript editing assistance from Tammy Markus, BA, and Dong Heon Lee, BA.
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Additional information: The e-Figure and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
FUNDING/SUPPORT: The Framingham Heart Study is funded by National Institutes of Health contract N01-HC-25195, HHSN268201500001I, and 75N92019D00031 (Boston University). This project was funded in part by the Division of Intramural Research, National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH), Bethesda, MD. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
Supplementary Data
References
- 1.Rubio D.M., Schoenbaum E.E., Lee L.S., et al. Defining translational research: implications for training. Acad Med. 2010;85:470–475. doi: 10.1097/ACM.0b013e3181ccd618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suhre K., Arnold M., Bhagwat A.M., et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho J.E., Lyass A., Courchesne P., et al. Protein biomarkers of cardiovascular disease and mortality in the community. J Am Heart Assoc. 2018;7(14) doi: 10.1161/JAHA.117.008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao C., Chen G., Song C., et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun. 2018;9:3268. doi: 10.1038/s41467-018-05512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith B.M., Austin J.H., Newell J.D., Jr., et al. Pulmonary emphysema subtypes on computed tomography: the MESA COPD study. Am J Med. 2014;127(94):e7–e23. doi: 10.1016/j.amjmed.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johns D.P., Walters J.A., Walters E.H. Diagnosis and early detection of COPD using spirometry. J Thorac Dis. 2014;6:1557–1569. doi: 10.3978/j.issn.2072-1439.2014.08.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manichaikul A., Hoffman E.A., Smolonska J., et al. Genome-wide study of percent emphysema on computed tomography in the general population: The Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am J Respir Crit Care Med. 2014;189:408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogushi F., Hubbard R.C., Vogelmeier C., Fells G.A., Crystal R.G. Risk factors for emphysema: cigarette smoking is associated with a reduction in the association rate constant of lung alpha 1-antitrypsin for neutrophil elastase. J Clin Invest. 1991;87:1060–1065. doi: 10.1172/JCI115066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yonchuk J.G., Silverman E.K., Bowler R.P., et al. Circulating soluble receptor for advanced glycation end products (sRAGE) as a biomarker of emphysema and the RAGE axis in the lung. Am J Respir Crit Care Med. 2015;192:785–792. doi: 10.1164/rccm.201501-0137PP. [DOI] [PubMed] [Google Scholar]
- 11.Cheng D.T., Kim D.K., Cockayne D.A., et al. Systemic soluble receptor for advanced glycation endproducts is a biomarker of emphysema and associated with AGER genetic variants in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188:948–957. doi: 10.1164/rccm.201302-0247OC. [DOI] [PubMed] [Google Scholar]
- 12.Sun W., Kechris K., Jacobson S., et al. Common genetic polymorphisms influence blood biomarker measurements in COPD. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobbs B.D., deJong K., Lamontagne M., et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49:426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho M.H., Castaldi P.J., Hersh C.P., et al. A genome-wide association study of emphysema and airway quantitative imaging phenotypes. Am J Respir Crit Care Med. 2015;192:559–569. doi: 10.1164/rccm.201501-0148OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.1000 Genomes Project Consortium. Auton A., Brooks L.D., et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shrine N., Guyatt A.L., Erzurumluoglu A.M., et al. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet. 2019;51:481–493. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giambartolomei C., Vukcevic D., Schadt E.E., et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S., Neale B., Todd-Brown K., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MR-Base. 2-sample Mendelian Randomisation. http://www.mrbase.org/. Accessed January 8, 2018.
- 20.Sakornsakolpat P., Dmitry Prokopenko, Maxime Lamontagne, et al. Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat Genet. 2019;51:494–505. doi: 10.1038/s41588-018-0342-2. Accessed January 8, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann M.A., Drury S., Hudson B.I., et al. RAGE and arthritis: the G82S polymorphism amplifies the inflammatory response. Genes Immun. 2002;3:123–135. doi: 10.1038/sj.gene.6363861. [DOI] [PubMed] [Google Scholar]
- 22.Walter R.E., Wilk J.B., Larson M.G., et al. Systemic inflammation and COPD: the Framingham Heart Study. Chest. 2008;133:19–25. doi: 10.1378/chest.07-0058. [DOI] [PubMed] [Google Scholar]
- 23.Araki T., Putman R.K., Hatabu H., et al. Development and progression of interstitial lung abnormalities in the Framingham Heart Study. Am J Respir Crit Care Med. 2016;194:1514–1522. doi: 10.1164/rccm.201512-2523OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka S., Yamashiro T., Washko G.R., Kurihara Y., Nakajima Y., Hatabu H. Quantitative CT assessment of chronic obstructive pulmonary disease. Radiographics. 2010;30:55–66. doi: 10.1148/rg.301095110. [DOI] [PubMed] [Google Scholar]
- 25.dupont N.C., Wang K., Wadhwa P.D., Culhane J.F., Nelson E.L. Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: determinations of a panel of nine cytokines in clinical sample culture supernatants. J Reprod Immunol. 2005;66:175–191. doi: 10.1016/j.jri.2005.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. ISBN 3-900051-07-0(2008).
- 27.Wolf L., Herr C., Niederstrasser J., Beisswenger C., Bals R. Receptor for advanced glycation endproducts (RAGE) maintains pulmonary structure and regulates the response to cigarette smoke. PLoS One. 2017;12 doi: 10.1371/journal.pone.0180092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faiz A., van den Berge M., Vermeulen C.J., Ten Hacken N.H.T., Guryev V., Pouwels S.D. AGER expression and alternative splicing in bronchial biopsies of smokers and never smokers. Respir Res. 2019;20:70. doi: 10.1186/s12931-019-1038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caraher E.J., Kwon S., Haider S.H., et al. Receptor for advanced glycation end-products and World Trade Center particulate induced lung function loss: a case-cohort study and murine model of acute particulate exposure. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H., Park J.-R., Kim W.J., et al. Blockade of RAGE ameliorates elastase-induced emphysema development and progression via RAGE-DAMP signaling. FASEB J. 2017;31:2076–2089. doi: 10.1096/fj.201601155R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen M., Wang T., Shen Y., et al. Knockout of RAGE ameliorates mainstream cigarette smoke-induced airway inflammation in mice. Int Immunopharmacol. 2017;50:230–235. doi: 10.1016/j.intimp.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi K., Iwamoto H., Horimasu Y., et al. AGER gene polymorphisms and soluble receptor for advanced glycation end product in patients with idiopathic pulmonary fibrosis. Respirology. 2017;22:965–971. doi: 10.1111/resp.12995. [DOI] [PubMed] [Google Scholar]
- 33.Oczypok E.A., Perkins T.N., Oury T.D. All the “RAGE” in lung disease: the receptor for advanced glycation endproducts (RAGE) is a major mediator of pulmonary inflammatory responses. Paediatr Respir Rev. 2017;23:40–49. doi: 10.1016/j.prrv.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miniati M., Monti S., Basta G., Cocci F., Fornai E., Bottai M. Soluble receptor for advanced glycation end products in COPD: relationship with emphysema and chronic cor pulmonale: a case-control study. Respir Res. 2011;12:37. doi: 10.1186/1465-9921-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalea A.Z., Schmidt A.M., Hudson B.I. Alternative splicing of RAGE: roles in biology and disease. Front Biosci (Landmark Ed) 2011;16:2756–2770. doi: 10.2741/3884. [DOI] [PubMed] [Google Scholar]
- 36.Gopal P., Reynaert N.L., Scheijen J.L.J.M., et al. Association of plasma sRAGE, but not esRAGE with lung function impairment in COPD. Respir Res. 2014;15:24. doi: 10.1186/1465-9921-15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhernakova D.V., Le T.H., Kurilshikov A., et al. Individual variations in cardiovascular-disease-related protein levels are driven by genetics and gut microbiome. Nat Genet. 2018;50:1524–1532. doi: 10.1038/s41588-018-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houghton A.M. Matrix metalloproteinases in destructive lung disease. Matrix Biol. 2015;44-46:167–174. doi: 10.1016/j.matbio.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Izushi Y., Kiyoshi Teshigawara, Keyue Liu, et al. Soluble form of the receptor for advanced glycation end-products attenuates inflammatory pathogenesis in a rat model of lipopolysaccharide-induced lung injury. J Pharmacol Sci. 2016;130:226–234. doi: 10.1016/j.jphs.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Blondonnet R., Audard J., Belville C., et al. RAGE inhibition reduces acute lung injury in mice. Sci Rep. 2017;7:7208. doi: 10.1038/s41598-017-07638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milutinovic P.S., Alcorn J.F., Englert J.M., Crum L.T., Oury T.D. The receptor for advanced glycation end products is a central mediator of asthma pathogenesis. Am J Pathol. 2012;181:1215–1225. doi: 10.1016/j.ajpath.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang F., Su X., Huang G., et al. sRAGE alleviates neutrophilic asthma by blocking HMGB1/RAGE signalling in airway dendritic cells. Sci Rep. 2017;7:14268. doi: 10.1038/s41598-017-14667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Robaiy S., Weber B., Simm A., et al. The receptor for advanced glycation end-products supports lung tissue biomechanics. Am J Physiol Lung Cell Mol Physiol. 2013;305:L491–L500. doi: 10.1152/ajplung.00090.2013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.