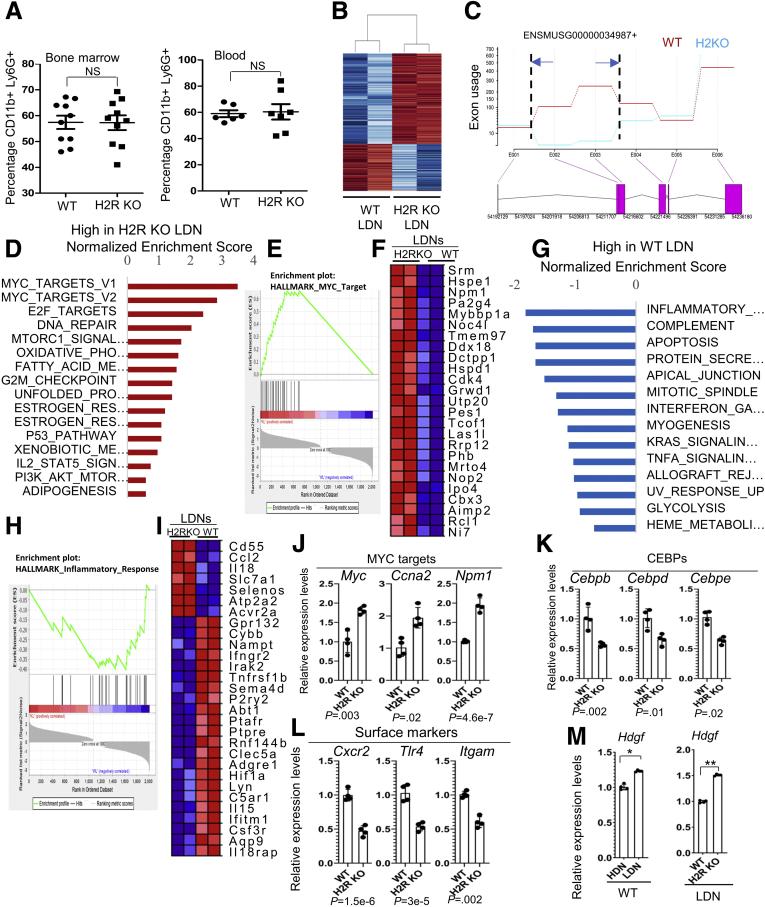
Figure 6.
H2R deficiency altered features of LDNs. (A) Neutrophil populations (CD11b+ Ly6g+) in bone marrow (n = 10) and in blood (n = 7) of WT and H2R-deficient mice. (B) A heat map of representative genes of WT and H2R-deficient LDNs. (C) The confirmation of HRH2 gene deletion (exons 2 and 3) by exon use by RNA sequencing. (D) Identification of the top pathways that are enriched most significantly in H2R KO LDNs were assessed by gene set enrichment analysis using Hallmark gene sets. (E and F) The enrichment plot and gene list of MYC targets in WT and H2R-deficient LDNs. (G) Identification of the top pathways that are enriched most significantly in WT LDNs were assessed by gene set enrichment analysis using Hallmark gene sets. (H and I) The enrichment plot and gene list of the inflammatory response pathway in WT and H2R-deficient LDNs. (J–L) The expression of representative genes in MYC target, CEBP family members, and surface markers of WT and H2R-deficient LDNs by qRT-PCR (n = 4). (M) Hdgf expression in HDNs and LDNs of WT and H2R-deficient mice (n = 4) by qRT-PCR. All values are means ± SD. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. CEBP, CCAAT/enhancer binding protein; KL, Knockout low-density; WL, WT low-density.