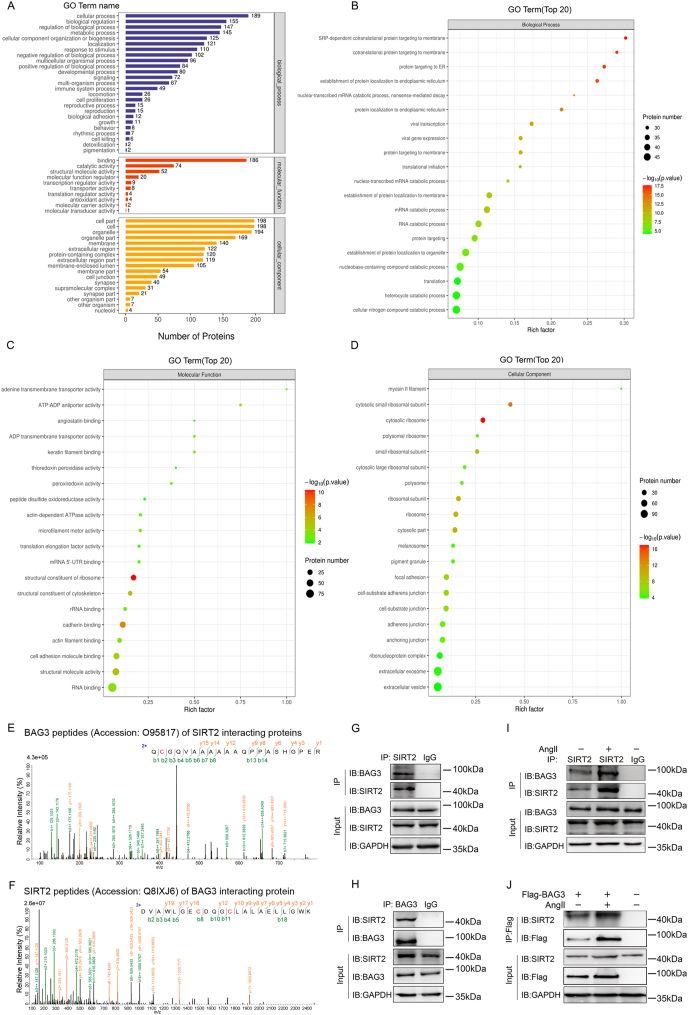
Fig. 5.
Mass spectrometry identifies BAG3 as a new SIRT2 substrate
(A) Mass spectrometry analysis to identify SIRT2-interacting proteins in HEK293T cells. The resulting SIRT2-interacting proteins were assessed by Gene Ontology enrichment analysis. The pathways with highest significance were related to protein binding. **P < 0.01, Fisher's exact test. (B-D) Mass spectrometry-detected SIRT2 interacting proteins in HEK293T cells were assessed for Biological Process, Molecular Function and Cellular Component enrichment analyses to screen physiological substrates of SIRT2. **P < 0.01, Fisher's exact test. (E) The spectrograms showed mass spectroscopy-identified BAG3 peptides (Accession: O95817; Description: BAG family molecular chaperone regulator 3 OS=Homo sapiens OX = 9606 GN = BAG3 PE = 1 SV = 3) of SIRT2 interacting proteins in HEK293T cells. (F) The spectrograms showed mass spectroscopy-identified SIRT2 peptides (Accession: Q8IXJ6; Description: NAD-dependent protein deacetylase sirtuin-2 OS=Homo sapiens OX = 9606 GN = SIRT2 PE = 1 S V = 2) of BAG3 interacting proteins in HEK293T cells. (G-H) Interaction of endogenous SIRT2 and BAG3 in HUVECs determined by immunoprecipitation with the indicated antibodies. (I) HUVECs underwent incubation without or with Ang II. Then, BAG3's interaction with SIRT2 was determined by immunoprecipitation with anti-SIRT2 antibodies, followed by immunoblot with anti-BAG3 antibodies. (J) Interaction between BAG3 and SIRT2 in presence or absence of Ang II determined by immunoprecipitation with anti-Flag-beads in HUVECs, followed by immunoblot with anti-SIRT2 antibodies.