FIGURE 1.
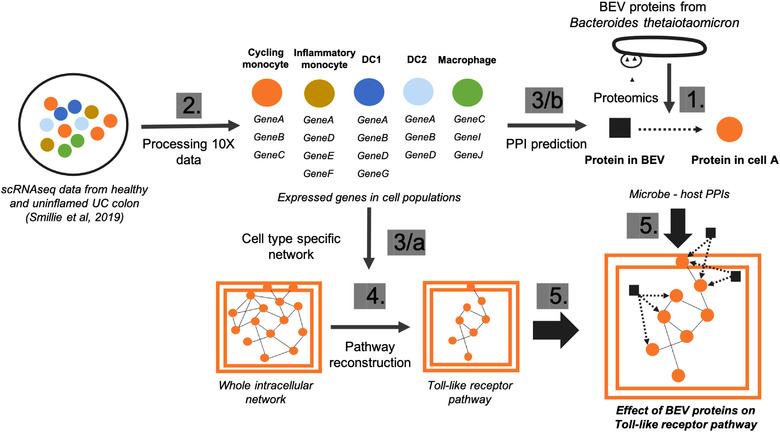
Computational workflow to analyse cell‐type specific effects of BEVs. Numbers indicate the sequence of the main steps: 1, Extraction of BEV proteins from the proteomic dataset 2, Processing the raw single‐cell transcriptomics from human colon 3/a, Creating cell‐type specific network using protein‐protein interactions from OmniPath (Türei et al., 2016) 3/b, Predicting protein‐protein interactions (PPIs) between BEV and host proteins in each cell‐type separately 4, Reconstruction of Toll‐like receptor pathway using Reactome database (Jassal et al., 2020) 5, Combining cell‐specific signalling with BEV targeted human proteins
