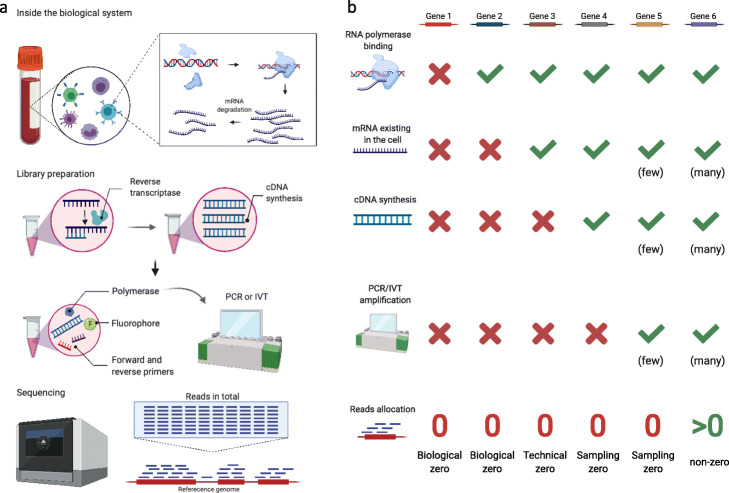
Fig. 1.
Sources of zeros in scRNA-seq data. a An overview of a scRNA-seq experiment. Biological factors that determine true gene expression levels include transcription and mRNA degradation (top panel). Technical procedures that affect gene expression measurements include cDNA synthesis, PCR or IVT amplification, and sequencing depth (bottom three panels). Finally, every gene’s expression measurement in each cell is defined as the number of reads or UMIs mapped to that gene in that cell. b How the biological factors and technical procedures in (a) lead to biological, technical, and sampling zeros in scRNA-seq data. Red crosses indicate occurrences of zeros, while green checkmarks indicate otherwise. Biological zeros arise from two scenarios: no transcription (gene 1) or no mRNA due to faster mRNA degradation than transcription (gene 2). If a gene has mRNAs in a cell, but its mRNAs are not captured by cDNA synthesis, the gene’s zero expression measurement is called a technical zero (gene 3). If a gene has cDNAs in the sequencing library, but its cDNAs are too few to be captured by sequencing, the gene’s zero expression measurement is called a sampling zero. Sampling zeros occur for two reasons: a gene’s cDNAs have few copies because they are not amplified by PCR or IVT (gene 4), or a gene’s mRNA copy number is too small so that its cDNAs still have few copies after amplification (gene 5). If the factors and procedures above do not result in few cDNAs of a gene in the sequencing library, the gene would have a non-zero measurement (gene 6). The figure is created with https://biorender.com/