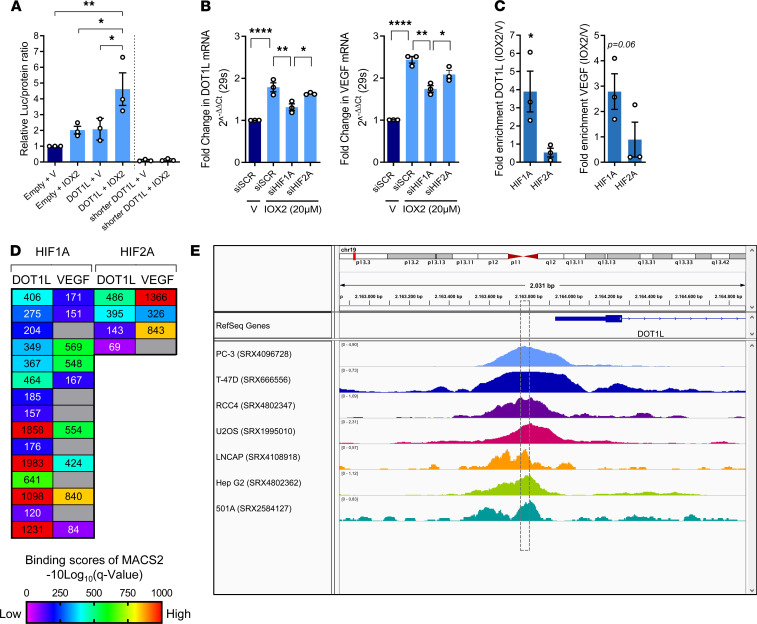
Figure 4. Hypoxia-mediated induction of DOT1L depends on HIF1A but not HIF2A.
(A) Luciferase assay in C28/I2 cells transfected with empty plasmid, full DOT1L promoter reporter, or negative control shorter DOT1L-promoter reporter, without conserved overlapping tandem HREs, upon treatment with IOX2 (20μM) for 72 hours, normalized to total protein relative to empty plasmid and vehicle (V) (n = 3, *P < 0.05, **P < 0.01, Šidák corrected for 6 tests in 2-way ANOVA). (B) Real-time PCR with siRNA-mediated silencing of HIF1A, HIF2A, or scrambled control (siSCR) (n = 3, *P < 0.05, **P < 0.01, ****P < 0.0001, Šidák corrected for 6 tests in 1-way ANOVA). (C) ChIP quantitative PCR (ChIP-qPCR) for HIF1A and HIF2A binding to DOT1L and VEGF promoters in cells treated with IOX2 (20 μM) for 72 hours (n = 3, *P < 0.05 by 1-sided t test). (D) MACS2-binding scores around DOT1L and VEGF transcription start site (TSS) of publicly available HIF1A and HIF2A ChIP-Seq data (ChIP-Atlas database). (E) Visualization of HIF1A ChIP-Seq in various cells around the DOT1L TSS. Box indicates overlapping HREs. ChIP-Atlas data mapped to reference human genome (hg19) using Integrative Genomics Viewer (IGV). Data are shown as the mean ± SEM.