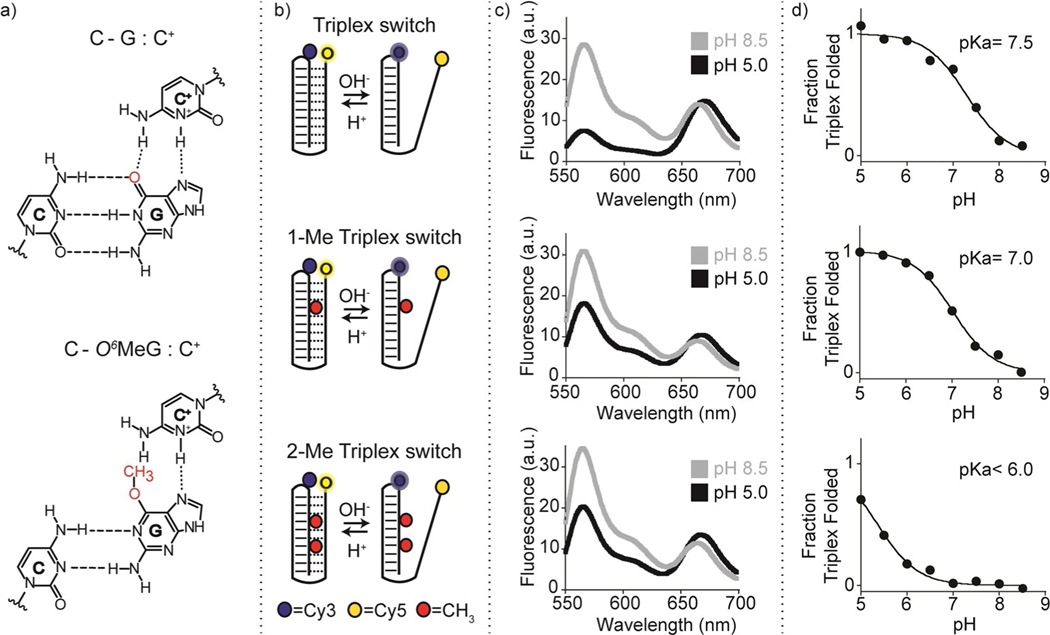
Figure 1.
Folding-upon-repair DNA nanoswitches. a) DNA parallel triplets formed between two cytosines and one guanine and involving Watson–Crick (three hydrogen bonds) and Hoogsteen interactions (two hydrogen bonds) require the protonation of the N3 of cytosine in the third strand (top, left) and thus are only stable at acidic pH values (average pKa of protonated cytosines in C-G:C triplet is ≈6.5). Because it affects Hoogsteen base pairing efficiency, methylation of the guanine in the O6-position destabilizes the triplex conformation. b) Programmable DNA-based triplex nanoswitches designed to form an intramolecular triplex structure with 0 (Triplex), 1 (1-Me Triplex) or 2 (2-Me Triplex) O6-MeG in the sequence. c) Fluorescence spectra were obtained for each nanoswitch at pH 5.0 and pH 8.5. d) pH-titration curves of the triplex nanoswitches. Triplex-to-duplex transition is monitored through a pH-insensitive FRET pair at the 3’-end (Cy5) and internally located (Cy3). The pH titration experiments were performed at 25°C, [nanoswitch]=50 nM by measuring the fluorescence signal at different pH values in 50 mM Na2HPO4, 150 mM NaCl buffer. Spectra were obtained by excitation at 530(±5) nm and acquisition between 545 and 700 nm (±10) nm.