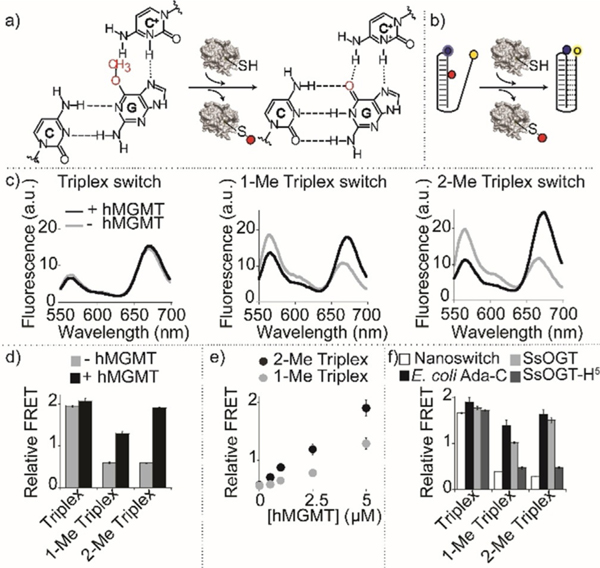
Figure 4.
Detection of methyltransferase activity with triplex nanoswitches. a) Methyltransferase enzymatic activity on DNA methylated nanoswitches leads to folding-upon-repair of the triplex DNA structure. b) Enzymatic activity detection can be achieved by monitoring folding/unfolding of the Triplex nanoswitch by fluorescence FRET signaling. c) Spectra and d) relative FRET signals obtained with the Triplex nanoswitches before and after incubation with 5 μM (0.1 μgμL−1) hMGMT. e) Relative FRET signals observed at different hMGMT concentrations for 1-Me and 2-Me Triplex nanoswitches. f) Relative FRET signals with different methyltransferase enzymes. Spectra were obtained by excitation at 530(±5) nm and acquisition between 545 and 700 nm (±10) nm. Relative FRET signals were obtained by first incubating the DNA nanoswitches (0.5 μM) in the absence or presence of AGTs (0.1 μgμL−1) in 10 μL solution of 50 mM Na2HPO4 buffer, 150 mM NaCl, pH 7.5 at 30°C for 60 minutes. The reaction mixtures were then diluted to 100 μL using 50 mM Na2HPO4 buffer, 250 mM NaCl at pH 5.0, and heat-inactivated for 2 minutes at 70°C before performing the fluorescence spectra at 25°C.