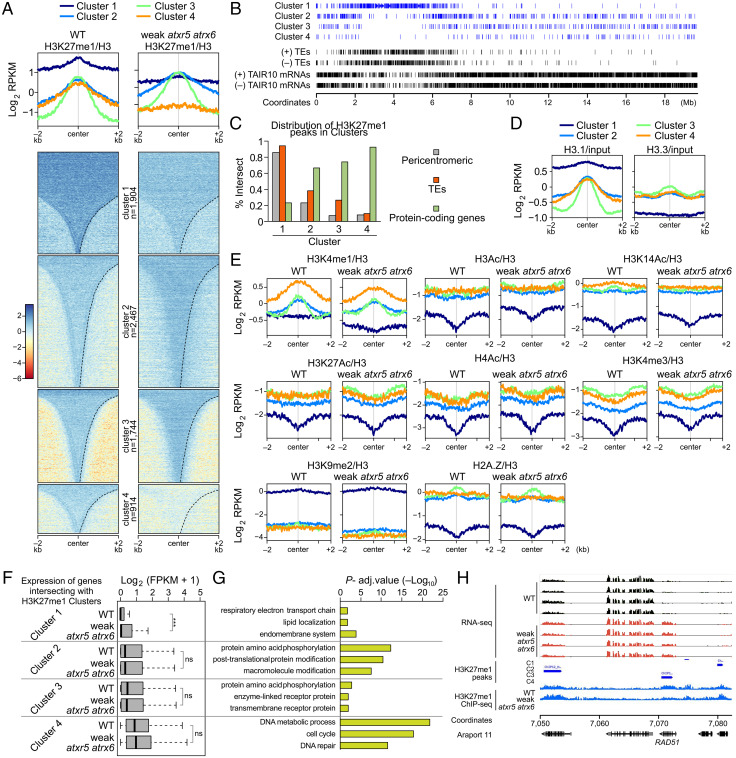
Fig. 2.
Characterization of chromatin in wild-type and atxr5/6 (W). (A) Profile and heatmap of normalized (log2 RPKM) H3K27me1 signal over H3K27me1-enriched peaks in wild type (MACS2, q < 0.01), grouped by k-means clustering (cluster n = 4) and ordered by region length. (B) Distribution of enriched regions for the four H3K27me1 clusters as defined in A over chromosome 2. (C) Overlap of regions in each of the four H3K27me1 clusters from A with pericentromeric regions (gray), TEs (red), and protein-coding genes (green). (D) Distribution of normalized (RPKM) H3.1 and H3.3 ChIP-seq signal (IP over input) in wild-type over H3K27me1 clusters (n = 4). Data are used with permission from Stroud et al. (49). (E) Distribution of H3K4me1, H3Ac, H3K14Ac, H3K27Ac, H4Ac, H3K4me3, H3K9me2, and H2A.Z and H3.3 ChIP-seq signal normalized to H3 in wild-type and atxr5/6 (W) cotyledons over the four H3K27me1 clusters. (F) Boxplot of RNA-seq log2 expression (average FPKM + 1) for genes overlapping each of the four H3K27me1 clusters in wild-type and atxr5/6 (W) cotyledons. Center lines indicate the median, upper and lower bounds represent the 75th and 25th percentiles, respectively, whiskers indicate the minimum and the maximum, and outliers are not plotted. n = 4 independent replicates in wild-type and atxr5/6 (W) cotyledons were averaged. Unpaired two-sample Wilcoxon test was used to test for significant differences in average expression in wild type vs. atxr5/6 (W) in each cluster: not significant (ns), P > 0.05; ***P ≤ 0.001. (G) GO term analysis for genes overlapping regions from each of the four H3K27me1 clusters. A −log adjusted P value is shown for three representative GO classes for each cluster. (H) Normalized H3K27me1 ChIP-seq signal (reads per kilobase of transcript per million mapped reads [RPKM]) and normalized RNA-seq signal (RPKM) in wild-type and atxr5/6 (W) cotyledons over RAD51. All RNA-seq tracks use same scale, as do both ChIP-seq tracks.