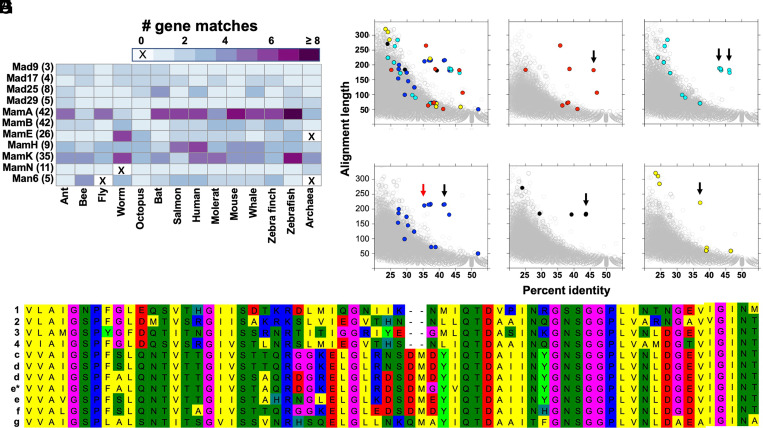
Fig. 2.
Comparative genomics. Data are presented for reciprocal BLASTp matches between magnetotactic bacterial biomineralization proteins and genome contents of eukaryotes and the archaea Lokiarchaeota. (A) Numbers of eukaryote proteins with reciprocal BLASTp match to 11 proteins known for involvement in prokaryote iron biomineralization (numbers of genes in prokaryote database in parenthesis). (B–G) Scatterplots of alignment lengths and percent identities scores for unidirectional BLASTp matches between genome contents of five magnetic responsive eukaryote taxa and the MTB magnetosome gene dataset (gray background circles). Proteins showing homology to the MTB gene MamE (HtrA-like serine protease) with E-value < 1 × 10e−5 are color highlighted. (B) All taxa (C–G combined), (C) zebra finch, Taeniopygia guttata (red); (D) naked mole-rat Heterocephalus glaber (cyan); (E) Chinook salmon, O. tshawytscha (blue); (F) little brown bat, Myotis lucifugus (black); and (G) honeybee, Apis mellifera (yellow). (H) A partial (66 amino acid) MamE alignment displays high levels of conservation across the five eukaryote taxa (C–G) and four MTB (1 to 4: UniprotKB accessions L0R6S4, Desulfamplus magnetovallimortis; C5JBP1, uncultured bacterium; A0A0F3GW16, Candidatus Magnetobacterium bavaricum; C5JAJ2, uncultured bacterium). Arrows in panels C to G point to the gene included in the multispecies alignment, with the red arrow indicating a gene differentially and more highly expressed in salmonid candidate magnetoreceptor cells, indicated by e* in the alignment. A full alignment is available from Dataset S4. Genome details are available from SI Appendix, Table S2.