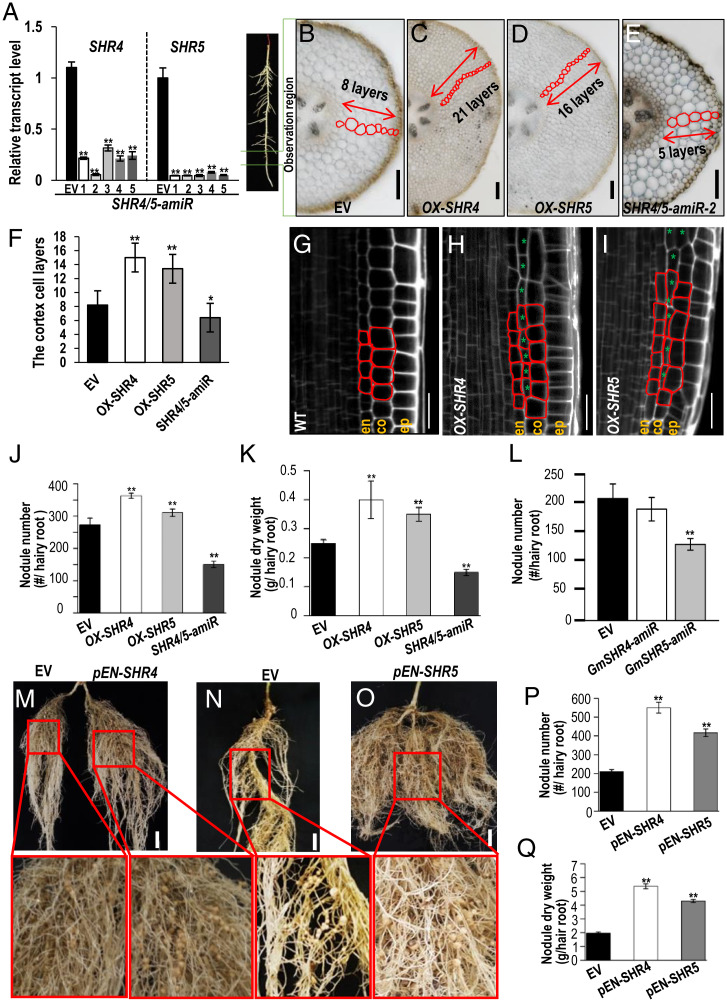
Fig. 2.
GmSHR4/5 expression level affects cortical cell division and root nodulation. (A) qRT-PCR shows that the expression of GmSHR4 and GmSHR5 is suppressed in the roots of artificial microRNA lines (GmSHR4/5-amiR). The RNA was extracted from the noninoculated roots. Error bars represent SD. GmEF1α (accession no. X56856) was used as the internal control. (B–E) The cortex cell layers in the root cross-section of GmSHR4/5 overexpression lines (OX-GmSHR4/5) and GmSHR4/5-amiR lines. All cortex sections were sampled from the same position of the hairy roots (n ≥ 15). (Scale bars, 100 μm.) (F) Quantification of the cortex cell layers shown in B–E (n ≥ 15). (G–I) Extra cortex cell division in the propidium iodide-stained Arabidopsis roots with ectopic expression of GmSHR4 and GmSHR5 (n ≥ 10). (Scale bars in G–I, 20 μm.) Green asterisks point to the extra cortex cells. (J and K) Nodule number (J) and nodule dry weight (K) in roots at 28 dai (n ≥ 25). (L) Nodule number per root at 28 DAI. (n ≥ 25). (M–Q) Nodulation in different backgrounds. (M) Split root showing the same root transformed with EV (Left) and pENOD40-GmSHR4 (Right). (N and O) EV and pENOD40-GmSHR5. Lower panels are the close-up views of the roots in M–O. (Scale bars, 2 cm.) (P) Nodule number. (Q) Nodule dry weight. EN, ENOD40 (n ≥ 20). Significant differences are observed between transgenic plants and EV plants. (**P < 0.01, *P < 0.05; Student’s t test). Error bars represent SD. n represents the number of independent biological samples. All experiments were repeated three times.