Fig. 3. Sex differences in placental transfer are driven by differences in bulk versus spike protein–specific Fc-glycan transfer.
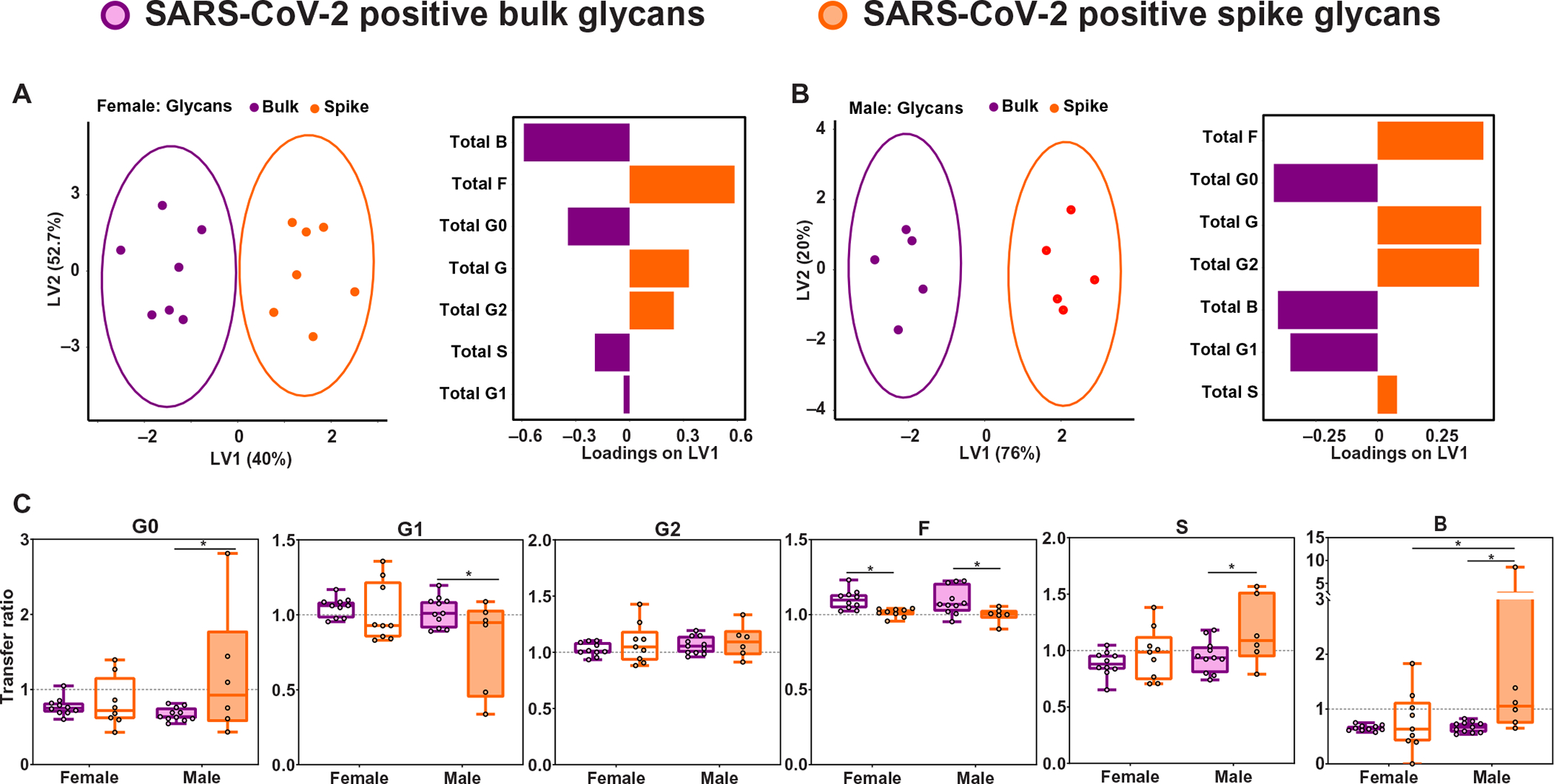
(A and B) A multilevel orthogonal partial least squares discriminant analysis (O-PLSDA) was built using bulk (purple) and spike protein–specific (orange) antibody glycan data from SARS-CoV-2–positive mothers pregnant with a female fetus (A) or male fetus (B) for which matched data were available. Each maternal sample is depicted as a dot on the scores plot (left). The glycan feature loadings on the first latent variable (LV1) are depicted by the bar graph (right). The model performance was assessed using leave-one-out cross-validation, with the average accuracy scores reported to be 98 and 100% predictive accuracy for the female and male O-PLSDA models, respectively. (C) Box and whisker plots showing the transfer ratios (cord:maternal ratios) for bulk and spike protein–specific Fc-glycans. Glycoforms depicted are agalactosylated (G0), monogalactosylated (G1), digalactyosylated (G2), fucosylated (F), sialylated (S), and bisected n-acetyl-glucosamine (GlcNAc, B). Bulk glycans are shown in purple, and spike protein–specific glycans are shown in orange (n = 8 to 11 per group). Pregnancies with female neonates are shown as open bars, whereas pregnancies with male neonates are shown as shaded bars. For box and whisker plots in (C), box extends from the 25th to 75th percentile, the whiskers depict minimum and maximum, and horizontal line depicts the median. Differences across groups were assessed by two-way ANOVA followed by FDR multiple comparisons correction post hoc analyses. *P < 0.05.
