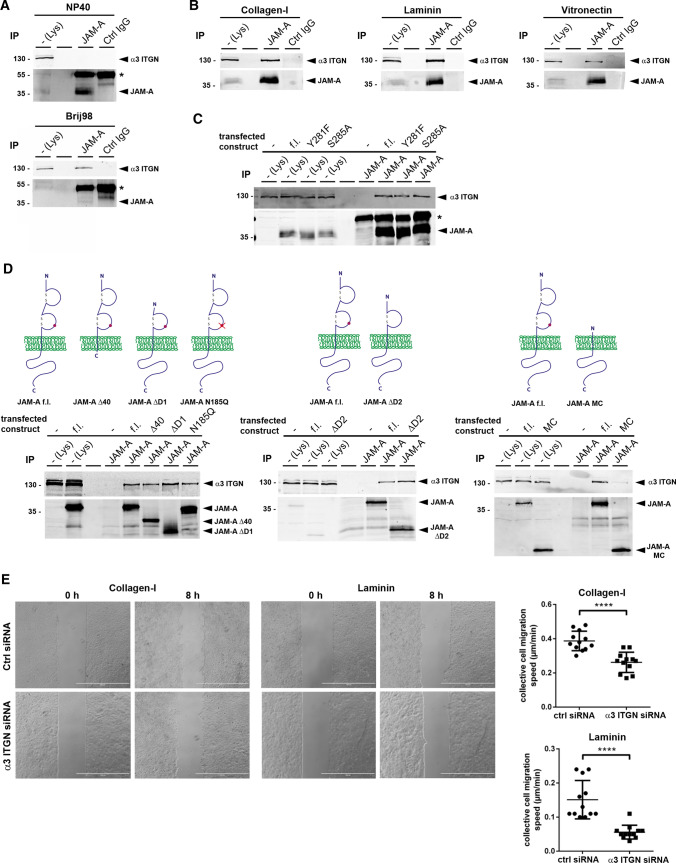
Fig. 4.
JAM-A interacts with α3β1 integrin in MDCKII cells. A CoIP of α3β1 integrin with JAM-A. The interaction of JAM-A with α3β1 integrin is detectable in Brij98-based lysates but not in NP40-based lysates. Note that the weak JAM-A signal in Brij98 lysed cells is due to lower solubility of JAM-A in Brij98. The asterisks indicate IgG heavy chains. IP immunoprecipitation, Lys lysate. B CoIP of α3β1 integrin with JAM-A from MDCKII cells cultured on collagen-I, laminin or vitronectin. JAM-A immunoprecipitates were immunoblotted for α3 integrin and JAM-A as indicated. To increase the solubility of JAM-A, the direct IPs for JAM-A were performed with NP40-based lysates. C CoIPs of JAM-A phospho-deficient mutants (JAM-A/Y281F, JAM-A/S285A) with α3β1 integrin. The asterisk indicates IgG heavy chains. D CoIPs of JAM-A mutants with α3β1 integrin. JAM-A constructs with deletions of the entire cytoplasmic domain (Δ40), the membrane-distal Ig-like domain (ΔD1), the membrane-proximal Ig-like domain (ΔD2), the entire extracellular domain (MC), or with point mutations affecting the N-linked glycosylation site of JAM-A (N185Q), were transfected into JAM-A-deficient MDCKII cells and analyzed for interaction with α3β1 integrin by CoIP. f.l. full length, MC membrane and cytoplasmic. E Collective cell migration velocity of control MDCKII cells (Ctrl siRNA pool) and α3 integrin KD MDCKII cells (α3 ITGN siRNA pool) on collagen-I and laminin. Left: representative images of monolayer expansion immediately after stamp removal (0 h) and after 8 h (8 h). Right: statistical evaluation of monolayer expansion on collagen-I and laminin. Each dot represents one independent cell population. Number of independent cell populations analyzed: n = 12 for each condition. Data are derived from three independent experiments. Statistical analysis shown in this figure was performed using two-tailed Student’s t test. Data are presented as mean values ± SD. ****P < 0.0001