Figure 3.
Cellular disruption and tissue identity loss from SARS-CoV-2 infection
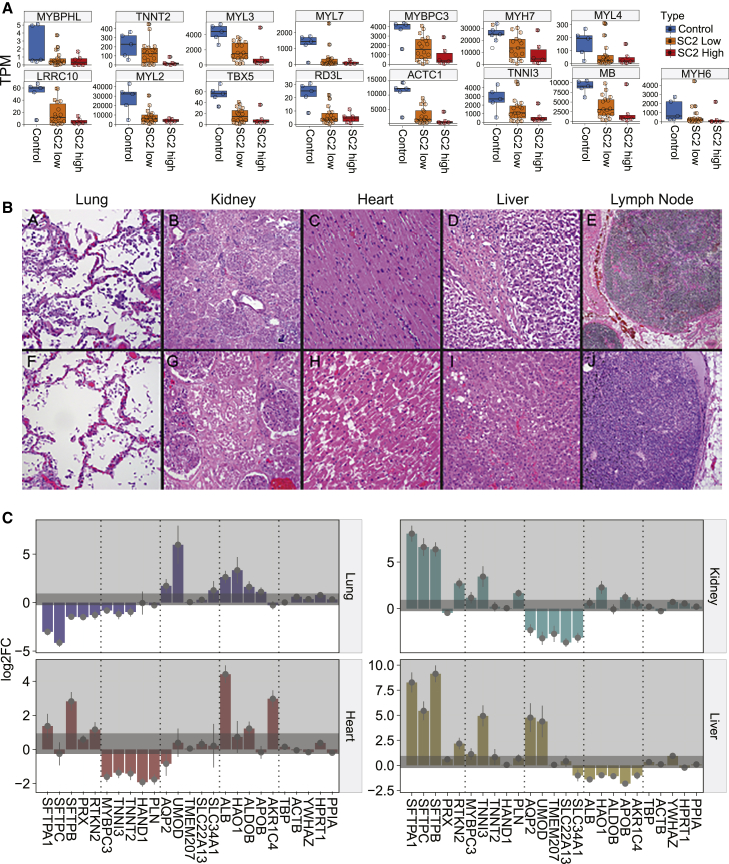
(A) Specific gene expression distributions for cardiomyocyte-related genes (from patient autopsy samples, n = 41).
(B) Sample collection strategy for COVID-19 autopsy samples from complete adult cases.
(A–J) Two cases with representative hematoxylin and eosin (H&E) images are shown: (A–E) Cases 58 and (F–J) 73. (A and F) Lung, (B and G) kidney, (C and H) heart, (D and I) liver, and (E and J) mediastinal lymph node (H&E stain). (B, D, E, I, and J) Original magnification ×50. (A, C, F, G, and H) Original magnification ×100.
(C) Gene expression changes of representative functional markers for each organ: heart (n = 41), kidney (n = 27), liver (n = 40), and lung (n = 40). Five genes related to the organ function were chosen (from the Human Protein Atlas), along with five housekeeping genes on the rightmost side. Darker gray shaded area represents the ranges of the housekeeping gene changes (baseline noise) and error bars show standard error from DESeq2 output, relative to the uninfected control.