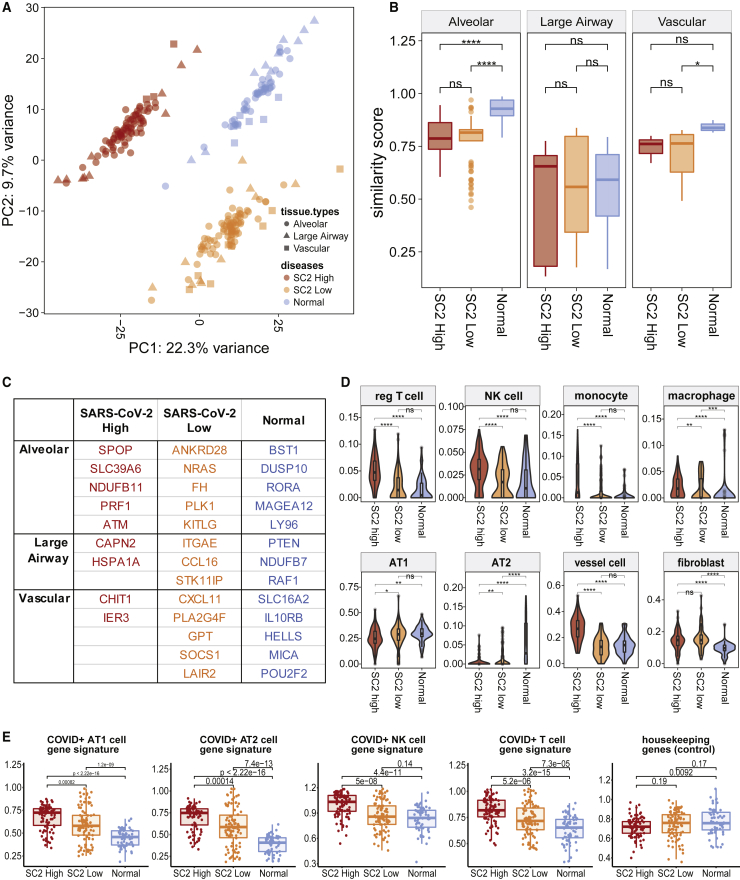
Figure 5.
Evaluation of tissue identity loss and heterogeneity from infection
(A) Principal-component analysis (PCA) to see sample- and disease-clustering. Colors denote disease conditions (64 normal ROIs, 86 and 97 SARS-CoV-2 high- and low-viral-load ROIs), while shapes show the tissue types (alveolar, large airway, and vascular regions).
(B) Similarity score distribution when compared with a tissue-specific (278 alveolar, 45 large airway, and 34 vascular ROIs) healthy reference gene profile (ns, non-significant, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001).
(C) Genes identifying disease- and tissue-specific conditions.
(D) Cell-type proportions in normal versus COVID-19. The median and quartiles are noted by the box plot inside. p value two-tailed t tests were done to compare the means (ns, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001).
(E) Enrichment of cell type- and COVID-19-specific gene signatures from 11 patients across 247 ROIs.