Extended Data Fig.2. Temporal transitions in nervous system gene expression across C. elegans post-embryonic development.
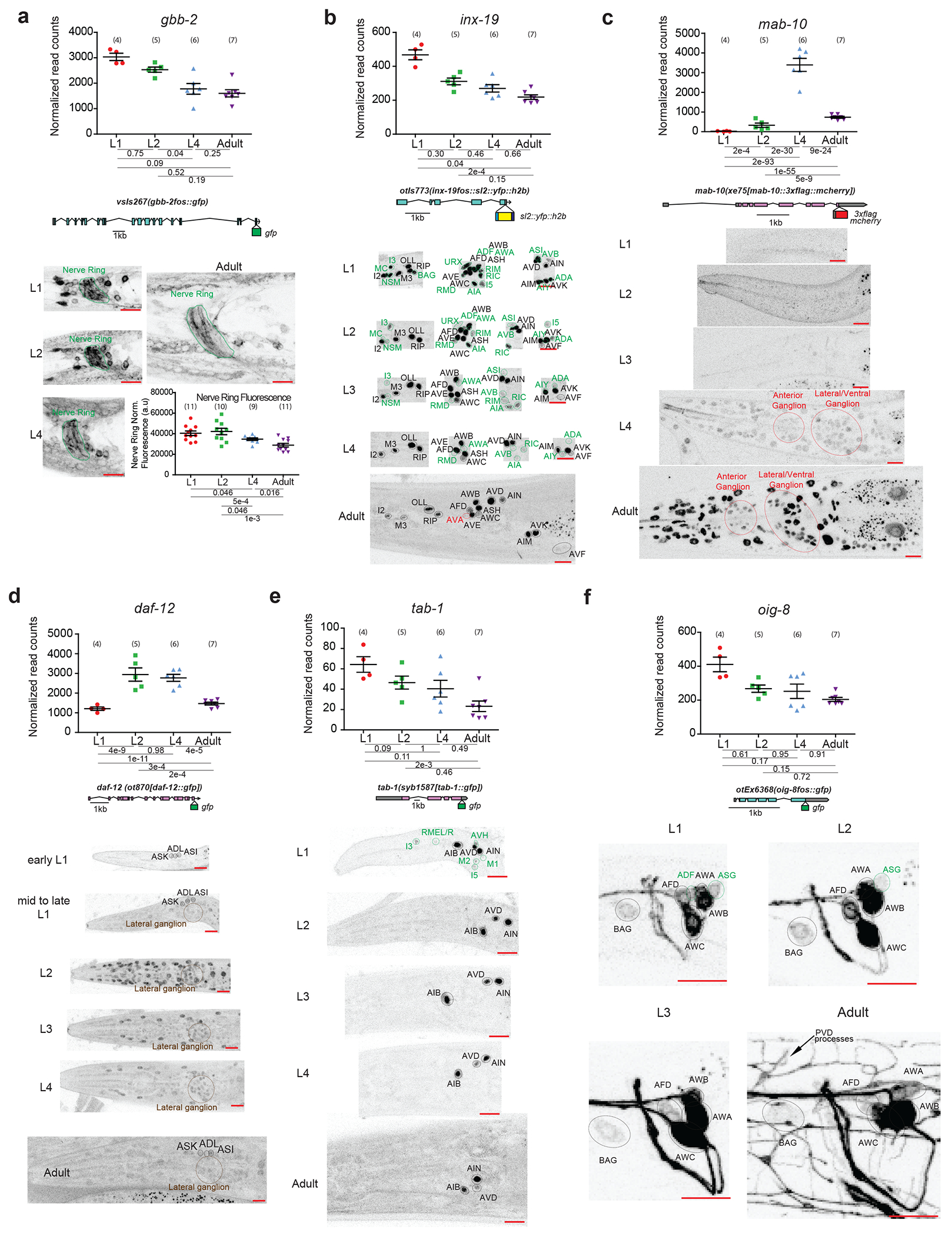
For all panels, validations of developmentally regulated genes with expression reporters are shown. On top are the scattered dot plots (each point represents a single replicate, n in bracket) of the normalized read counts across all developmental stages from the neuronal INTACT/RNA-seq profiling. Mean +/− SEM are shown for each stage. Adjusted p values (padj), as calculated by DESeq2, for each developmental comparison are below. Below the RNA-seq read count plots are the schematics and allele names of the expression reporters. Below that are representative confocal microscopy images of the expression reporters across development. Specific regions/neurons are labeled with dotted lines: those labelled with black dotted lines/names are not altered developmentally while those labelled with green and red lines/names demonstrate, respectively, decreases and increases in expression across development. Those labelled with brown lines/names demonstrate both increases and decreases in expression in the same neurons across development. Red scale bars (10μm) are on the bottom right of all representative images. For all panels, L1 through L4 represent the first through the fourth larval stage animals. For a, additional quantification of fluorescence intensity is also shown at the bottom. Post-hoc two-sided t-test p values and n (in bracket) are shown. Additional details are included in Supplementary Table 6.
a, Metabotropic glutamate receptor gbb-2, as validated with a translational fosmid reporter (gfp), shows expression in the same set of neurons across development 30, although the intensity of expression is decreased across development, including that in the nerve ring as measured with fluorescence intensity. b, Gap junction molecule inx-19, as validated with a transcriptional fosmid reporter (sl2::yfp::h2b), loses expression in sixteen neuronal classes across development and gains expression in the AVA neuron upon entry into adulthood.
c, Transcription cofactor mab-10, as validated with an endogenous translational reporter (3xflag::mcherry) engineered with CRISPR/Cas9, gains expression across the nervous system amongst other tissue during transition into the L4 stage that is further upregulated in adulthood. The RNA prediction matches well with previous RNA FISH analysis 52. The difference between RNA data and protein reporter expression is consistent with previous characterized post-transcriptional regulation by LIN-41 53. d, Nuclear hormone receptor daf-12, as validated with an endogenous translational reporter (gfp) engineered with CRISPR-Cas9, shows increased expression broadly across the nervous system during early/mid-larval stage and then decreased expression upon transition into late larval/adult stage. e, Homeodoman transcription factor tab-1, as validated with an endogenous translational reporter (gfp) engineered with CRISPR-Cas9, loses expression in five classes of neuron during early larval development. f, Immunoglobulin-like domain molecule oig-8, as validated with a translational fosmid reporter (gfp), loses expression in two classes of neuron during early larval development.
