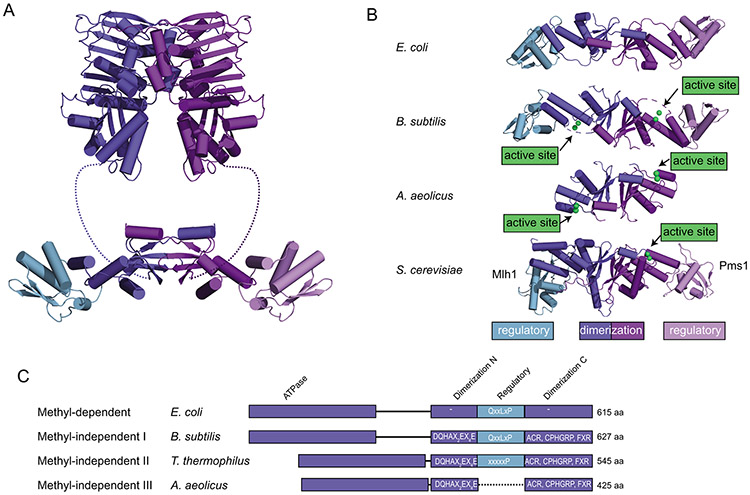
Figure 4. Features of the C-terminal domains of MutL homologs.
A. Ribbon diagram of E. coli MutL homolog comprised of N-terminal ATPase domains (top; PDB ID 1b62 (98)) and a C-terminal dimerization domain made up of a dimerization/nuclease subdomain and a regulatory subdomain (bottom; PDB ID 1x9z (99)). B. Comparison of the C-terminal domain structures of E. coli MutL (a methyl-directed MutL without an endonuclease active site; PDB ID 1x9z (99)), B. subtilis MutL (a methyl-independent MutL with an endonuclease active site; PDB ID 3kdk (64)), Aquifex aeolicus MutL (containing an endonuclease active site but not a regulatory subdomain; PDB ID 5b42 (151)), and S. cerevisiae Mlh1-Pms1 (containing an endonuclease active site and a regulatory subdomain in the Pms1 protein; PDB ID 4e4w (154)). C. Diagram of MutLs from methyl-dependent and methyl-independent subfamilies I-III. Endonuclease domains are missing from the methyl-dependent MutL proteins, but present in the methyl-independent MutLs. The methyl-independent subfamilies II and III lack the regulatory domain involved in interaction with the β-clamp through mutation (subfamily II) or deletion (subfamily III).