Figure 4.
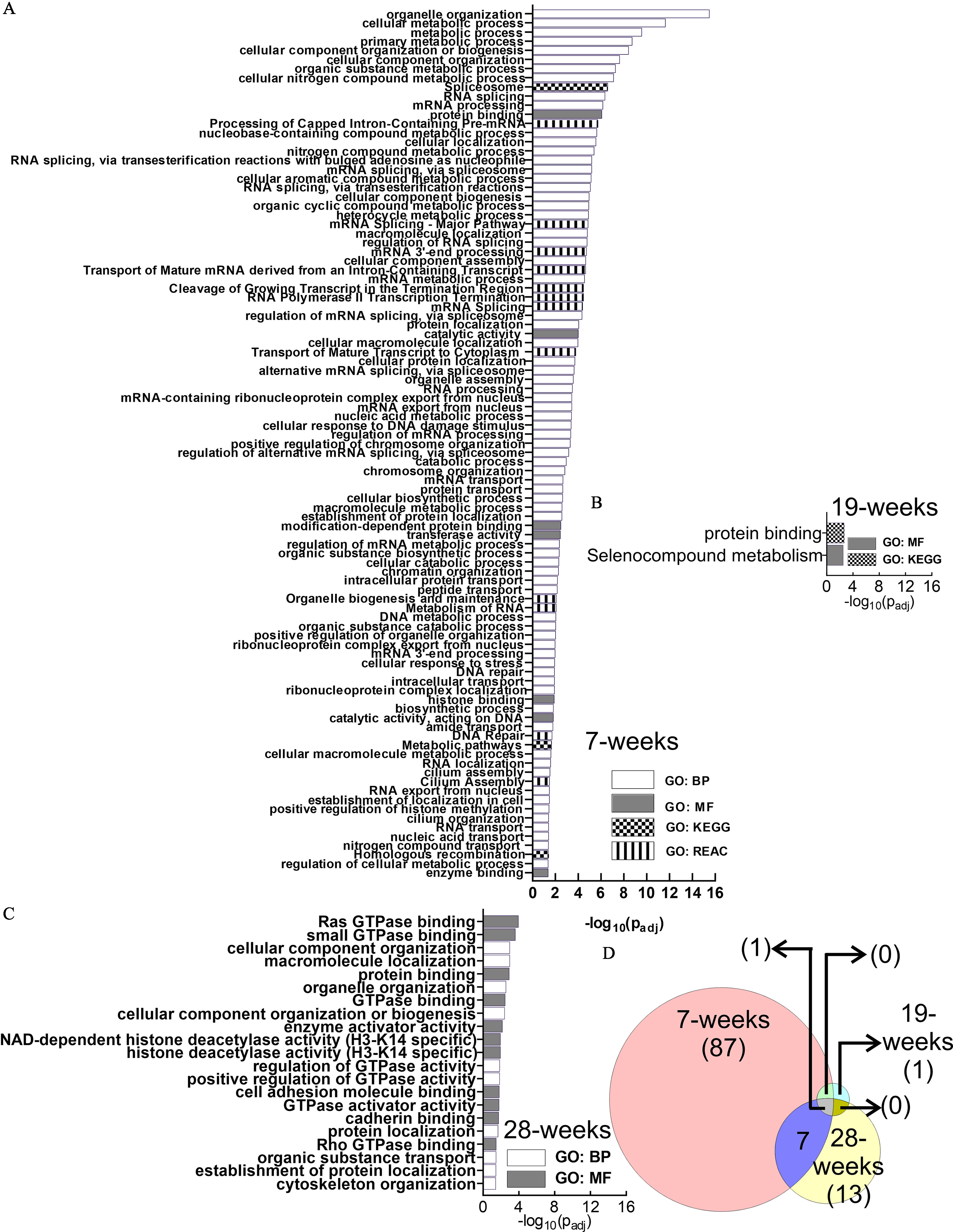
GO Functional enrichment analysis of the differential alternative splicing events in HaCaT cells induced by arsenic exposure. (A) 7 wk, (B) 19 wk, and (C) 28 wk. GO terms depicted in (A–C) are presented in the same order as in Excel Table S1 [in ascending order of ]. (D) Venn diagram describing the number of enriched pathways at each time point and their overlap at different time points. Note: BP, biological process; GO, gene ontology; GTPase, guanosine triphosphatase; KEGG, Kyoto Encyclopedia of Genes and Genomes; MF, molecular function; REAC, Reactome.
