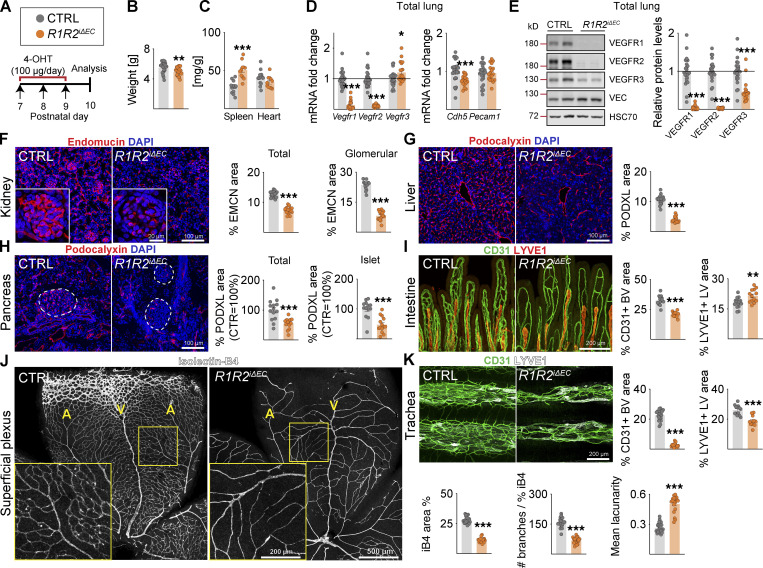
Figure 2.
VEGFR1 deletion aggravates vessel regression induced by VEGFR2 loss. (A) Experimental setup. (B) Pup weights on P10. (C) Spleen and heart weights normalized to body weight. (D) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (E) Western blots and quantifications of VEGFRs normalized to HSC70 and VE-Cadherin (VEC) in lung lysates. (F) Staining and quantification of blood vessels (EMCN, red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (G) Staining and quantification of blood vessels (PDXL, red) in the liver and (H) pancreas; dashed lines mark Langerhans islets. (I) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification. (J) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A, arteries; V, veins. Only the superficial plexus is shown. Scale bar = 500 µm, inset = 200 µm. (K) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white). EMCN, endomucin; PODXL, podocalyxin. Error bars = mean ± SEM (n = control [CTRL] = 21, R1R2iΔEC = 17 mice pooled from five independent experiments were used for the data shown in the panels). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData F2.