FIGURE 2.
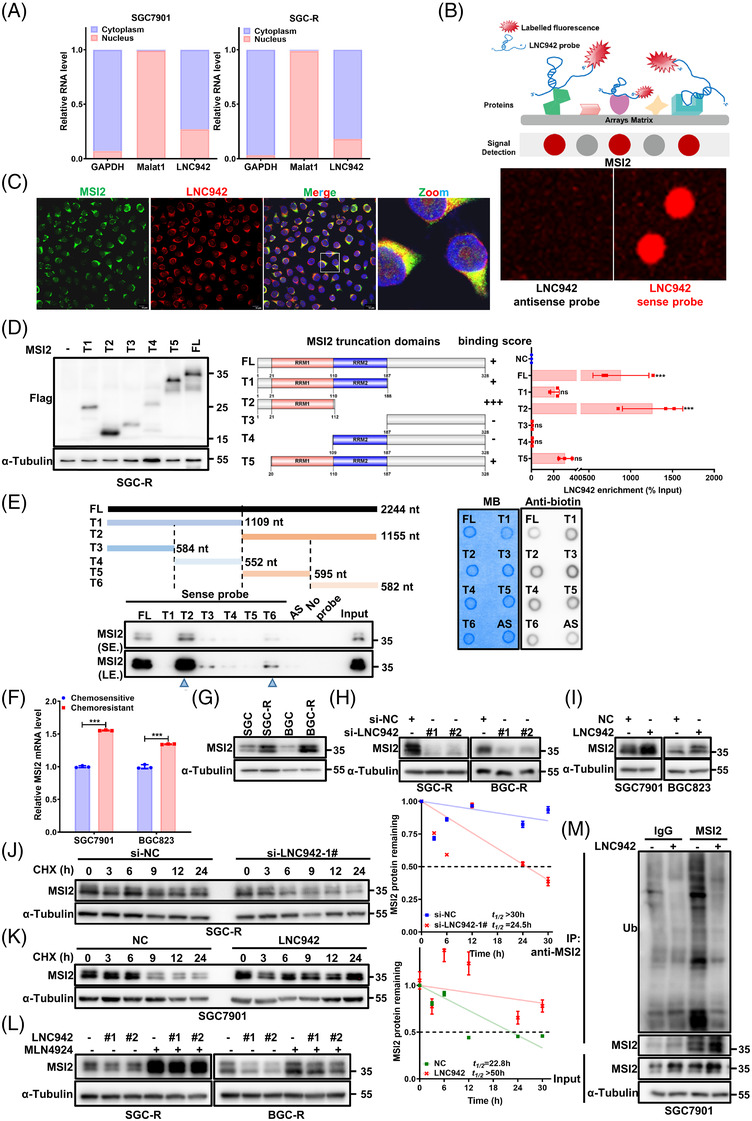
LNC942 interacts with MSI2 protein and prevents its ubiquitination. (A) Quantitative real‐time PCR (qRT‐PCR) detection of LNC942 expression in the cytoplasmic and nuclear fractions. GAPDH is the cytoplasmic marker, and Marat1 is the nuclear marker. (B) Identification of LNC942‐binding protein by human proteome microarray experiment. Upper: Schematic of the human proteome microarray experiment. Bottom: Protein spot subarray demonstrating strand‐specific binding of LNC942 sense strand to MSI2 protein. (C) RNA fluorescence in situ hybridization (FISH) and immunofluorescence (IF) assays showing the co‐localization of LNC942 (Cy3‐labelled, red) and MSI2 (Alexa488‐labelled, green) in SGC‐R cells. Blue: DAPI. All scale bars: 10 μm. (D) RNA immunoprecipitation (RIP) assays were performed with anti‐Flag antibody in SGC‐R cell transfected with vectors expressing Flag‐tagged full length (FL) or truncation mutants of MSI2. Left: Western blotting indicating the expression of Flag‐tagged FL or truncation mutants of MSI2. Middle: Schematic structures of MSI2 proteins and five truncated mutants of MSI2 variants. The binding score indicated the binding intensity between LNC942 and the domains of MSI2. Right: qRT‐PCR detection of LNC942 enrichment after RIP assays. (E) Deletion mapping of the MSI2‐binding region(s) in LINC00942. Left: Western blotting detecting the MSI2 enrichment after RNA pull‐down. AS: anti‐sense probe. Right: the biotin‐labelled LNC942 probes were quantified (300 ng) and detected by the dot blot assay. MB: methylene blue, used as internal reference. (F and G) qRT‐PCR (F) and western blotting (G) were performed for the detection of MSI2 expression in chemosensitive or resistant SGC7901 and BGC823 cells. (H) The protein levels of MSI2 in chemoresistant cells transfected with si‐NC or LNC942 siRNAs. (I) The MSI2 protein expression in SGC7901 and BGC823 cells stably expressing LNC942. (J) SGC‐R cells with or without LNC942 knock‐down were treated with cycloheximide (CHX) (100 μg/ml) for the indicated time points. MSI2 protein abundance was analysed through western blotting, followed by quantification using ImageJ. (K) Western blotting detection of MSI2 protein half‐life in LNC942 stably expressed SGC7901 cells with CHX (100 μg/ml) for the indicated time points. MSI2 protein abundance was quantified using ImageJ. (L) Western blotting of MSI2 in SGC‐R or BGC‐R cells transfected with or without si‐LNC942 following proteasome inhibition with MLN4924 (1 μM, 24 h). (M) SGC7901 cells stably expressing LNC942 were treated with 20 μM MG‐132 for 6 h and then subjected to IP assays to detect the MSI2 ubiquitination levels. Data in panels (A), (D), (F), (J) and (K) are represented as mean ± SD of the three independent experiments. The p value was determined using a two‐tailed unpaired Student's t test. ns, p > .05; *p < .05; **p < .01; ***p < .001
