Figure 2.
Macrophage DNA sensing induces cell communication in vitro
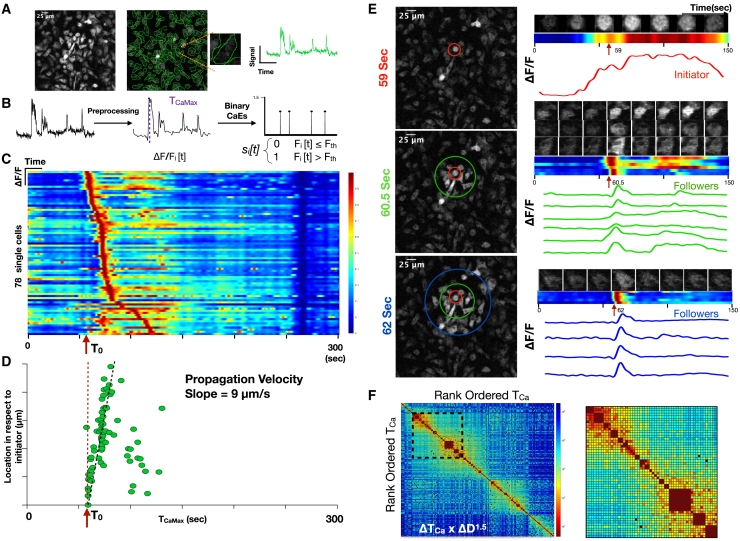
(A) Maximum image projection of BMDMs derived from the Csf1r-GCaMP5 calcium reporter mouse (left). ROIs are drawn to define cell boundaries (middle). Individual cell with corresponding single-cell fluorescence versus time tracing sampled at 2 Hz (right).
(B) Signal processing, including background correction, calcium intensity change quantification, bandpass filtering, peak-finding, and determination of time of maximum calcium elevation (TOCaEMax_CaEs) based on the magnitude of calcium intensity and converting the fluorescence peaks into impulses at the time of calcium elevation (TOCaE). Signal processing to define calcium elevation times and convert to a binary impulse train.
(C) Heatmap of single cells (y axis) versus time where intensity represents normalized change in fluorescence ΔF/F. The time of the initiating cell calcium elevation is defined as T0.
(D) Distance of cell from the initiating cell versus time of maximum calcium elevation intensity. This reveals a slope of 9 μm/s, which is interpreted as the communication propagation velocity.
(E) Frames of the time-lapse fluorescence during the propagation are shown at 59 s, 60.5 s, and 62 s (left). Color-coded concentric rings are shown to define the initiator cell, exhibiting sustained fluorescence (red), and the secondary responders (green and blue concentric rings), exhibiting brief calcium elevations. At right, time-lapse montage of individual cell calcium elevations are shown with the corresponding heatmap and fluorescence versus time tracing.
(F) Cross-correlation heatmap of rank-ordered calcium elevations for all cells. Color represents the product of calcium elevation time difference and Euclidean spatial distance ΔTOCaE × ΔD1.5. Inset shows the region of high cross-correlation indicating calcium elevations that are highly localized in time and space.