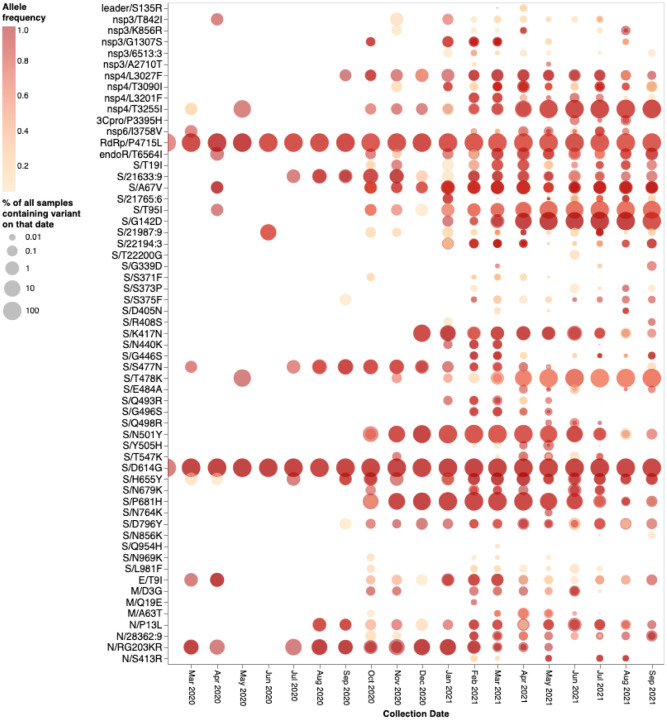
Figure 4. Intrapatient allelic variation seen at BA.1 amino acid mutation sites in a subset of SARS-CoV-2 raw sequencing data since March 2020 analyzed using a standardized variant calling pipeline24.
The areas of the circles indicate the proportions of raw sequence datasets (per 1,000 samples) where a mutation away from the Wuhan-Hu-1 consensus sequence was called. The colour of the circle indicates the median intrapatient allele frequency (AF) in datasets for which each mutation was detected. Mutations occurring at lower AFs are only present in a subpopulation of viruses in a particular host. The data has been generated by calling variants from read-level data of 230,506 samples from COG-UK, Estonia, Greece, Ireland, and South Africa: PRJEB37886, PRJEB42961 (and multiple other bioprojects with the study title: Whole genome sequencing of SARS-CoV-2 from Covid-19 patients from Estonia), PRJEB44141, PRJEB40277 and PRJNA636748. Note that S371L is the result of two nucleotide substitutions in codon S/371 and was never detected in intrapatient samples. S371F represents an intermediate mutation between the Wuhan-Hu-1 state and that of BA.1 and is presented here for completeness.