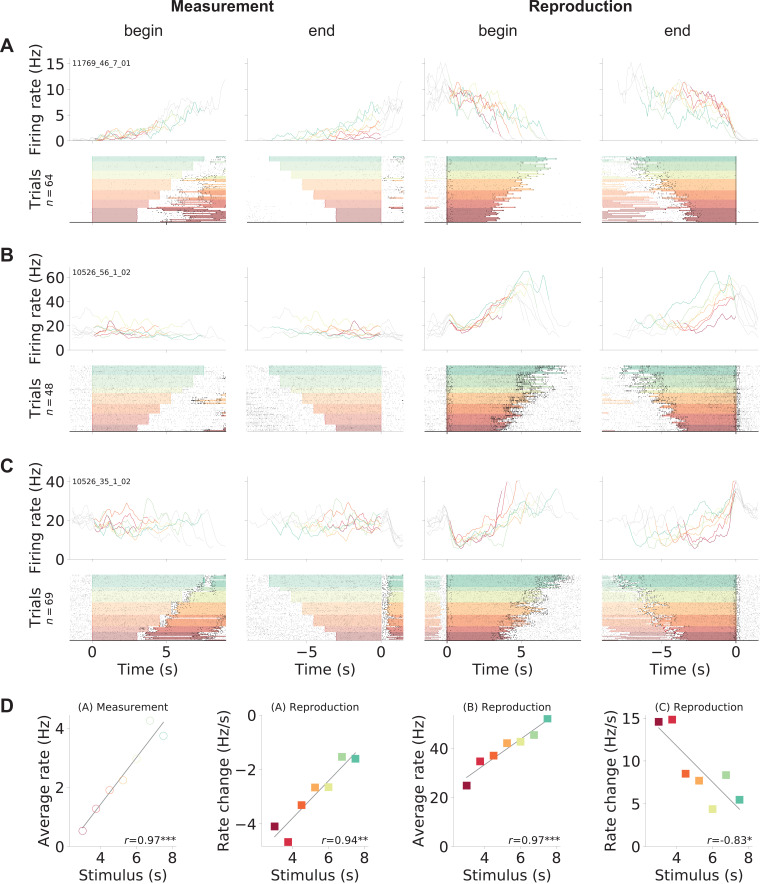
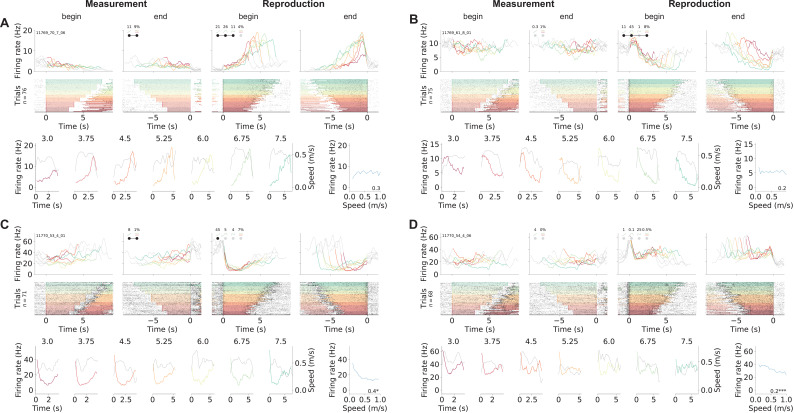
Figure 2. Gerbil medial prefrontal cortex (mPFC) neuron responses during time reproduction.
(A) A cell that linearly increased its firing rate during measurement and ramped down to zero during reproduction. (B) A neuron that scaled its firing with the stimulus duration during reproduction and (C) a ramp-to-threshold cell. (A–C) Panels display spike raster plots sorted by stimulus duration (bottom) and corresponding spike density functions (SDF, top). Each column plots the data with different alignment, that is, measurement begin and end, reproduction begin and end. Color identifies stimulus duration as in Figure 1C. In the raster plots, black ticks are single spikes. For better visualization, we only plot half of the spikes (randomly chosen). Measurement or reproduction phases are delimited by underlayed color. The SDFs are colored in the respective task phase, outside they are displayed as thin gray lines. (D) Dependence of firing on stimulus duration in the example cells. Single markers show the average firing rate or change of firing rate at each stimulus duration. Open dots are used for data from measurement and filled squares for those from reproduction. Solid lines are linear fits. Pearson’s correlation coefficient and significance is given in the lower-right corner. Above each panel, cell and task phase is indicated. The averages of firing rate and its change were calculated from the last half of the SDFs in the corresponding task phase.