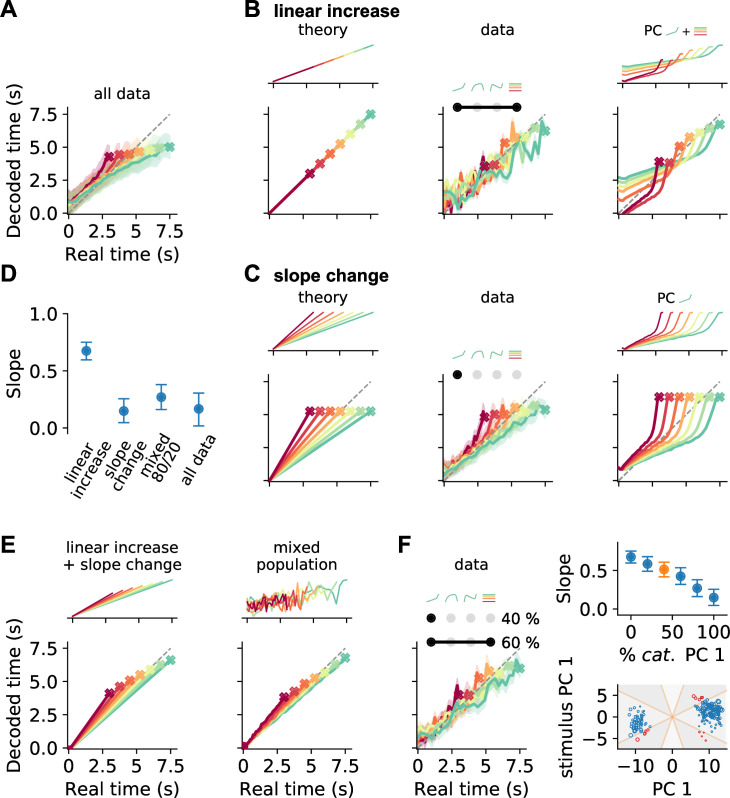
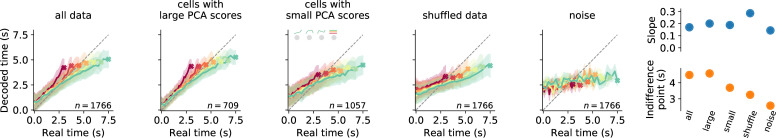
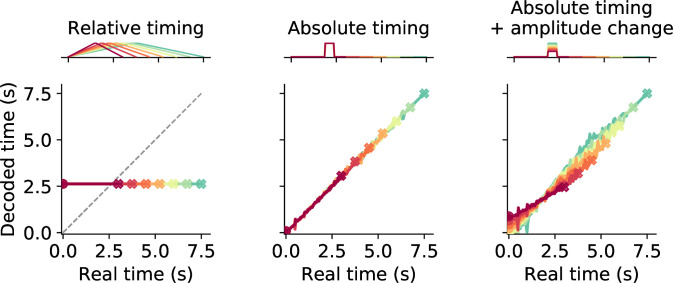
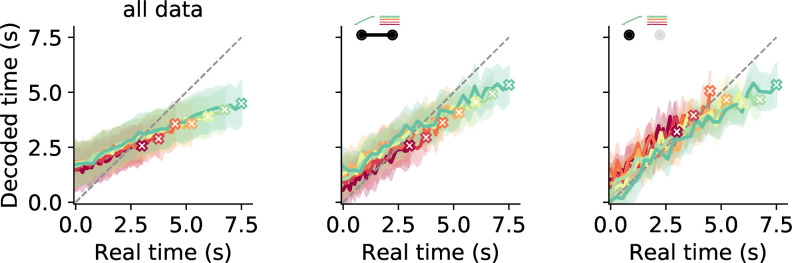
Figure 7. Decoding elapsed time from prefrontal population activity.
(A) Elapsed time (average ± standard deviation from bootstrapping) decoded from the responses of all neurons recorded during the reproduction phase. As before, color identifies stimulus duration. Crosses mark final values. A strong regression effect is visible. (B) Time decoding with neurons that ramp to stimulus-dependent levels but with the same slope and (C) with ramp-to-threshold cells. Left panels illustrate the theoretical prediction for decoded time and example neuronal activity above. Middle panels plot decoding results using only neurons from the respective response category. Right panels give results for decoding using the corresponding components from demixed principal component analysis (PCA). As displayed in the top panels, these components are principal component (PC) 1 and stimulus PC 1 in (B); and PC 1 only in (C). (D) Median slopes of the linear regression between the final values of real and predicted time for decoding from data in (A–C) and for a mixture of 80% slope-changing and 20% linear increasing cells, which aligns well with decoding from all data (A); regarding mixtures see also (F). Error bars delimit interquartile ranges (from bootstrapping). (E) Mixtures of linear increasing activity and slope changes explain behavioral regression effects. Theoretical predictions for mixing both responses in single neurons (left) or as two different response types across a population (right). For the second case, a neuron with noisy linear increasing activity is displayed as an example. (F) Decoding results for a mixed population of 40% slope-changing and 60% linear increasing cells. The cells were sampled at these fractions in each bootstrapping run from the response categories we identified in our recorded data. The upper-right panel shows the regression slope (D) for different fractions of slope-changing cells. The orange marker corresponds to the example in the left panel. The lower-right panel displays the PCA scores of the cells from the linear increasing (red; PC 1 + stimulus PC 1) and slope-changing categories (blue; PC 1 only). The size of the marker illustrates the decoder weight for that cell. See also Figure 5—figure supplement 4C.