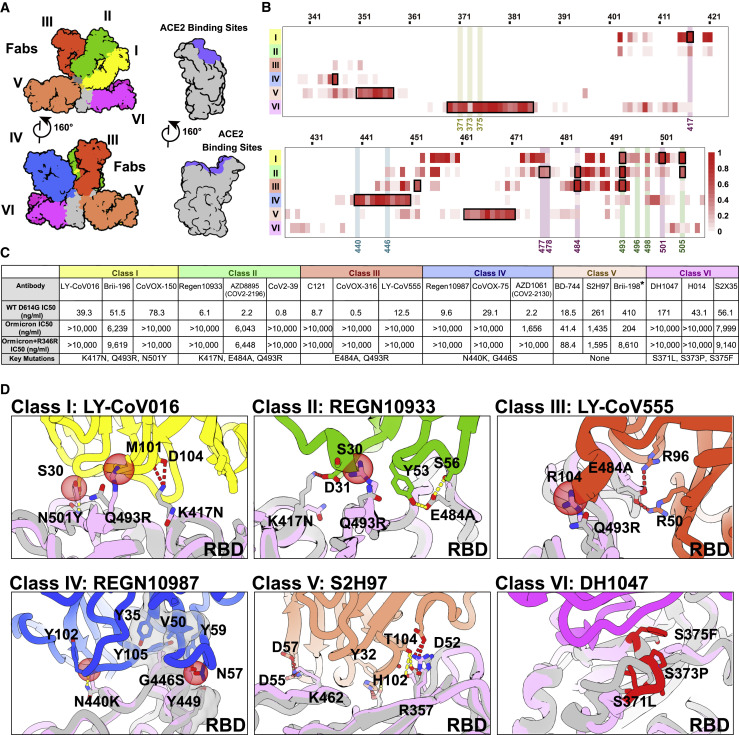
Figure 5.
Structural dissection of the evasion of neutralization of antibodies
(A) Surface representation of RBD in complex with six types of NAbs. RBD is colored in gray and the six representative Fab fragments belonging to six classes are colored as follows: class I, yellow; class II, green; class III, red; class IV, blue; class V, brown; class VI, magenta.
(B) Heatmap represents the frequency of RBD residues recognized by NAbs from six classes. Mutations present in Omicron RBD are marked out and highlighted.
(C) Summary of representative NAbs from each of six classes. Neutralizing titer (IC50) of each NAb against WT and Omicron is enumerated. The key residues involved in immune evasion for each class are also listed below. The IC50 data of Brii-198 was marked with “∗,” which represents the data referred from the available publication (Liu et al., 2021b). All neutralization assays were performed in biological triplicates.
(D) Binding interface between RBD and representative NAbs. All structures are shown as ribbon with the key residues shown with sticks. The clashes between RBD and NAb are shown as red sphere; salt bridges and hydrogen bonds are presented as red dashed lines and yellow dashed lines, respectively. Fab fragments of LY-CoV016, REGN10933, LY-CoV555, REGN10987, S2H97, and DH1047, representatives of classes I, II, III, IV, V, and VI, respectively, are colored according to the class they belong to; WT RBD is colored in gray; Omicron RBD is colored in light purple.
See also Figure S5.