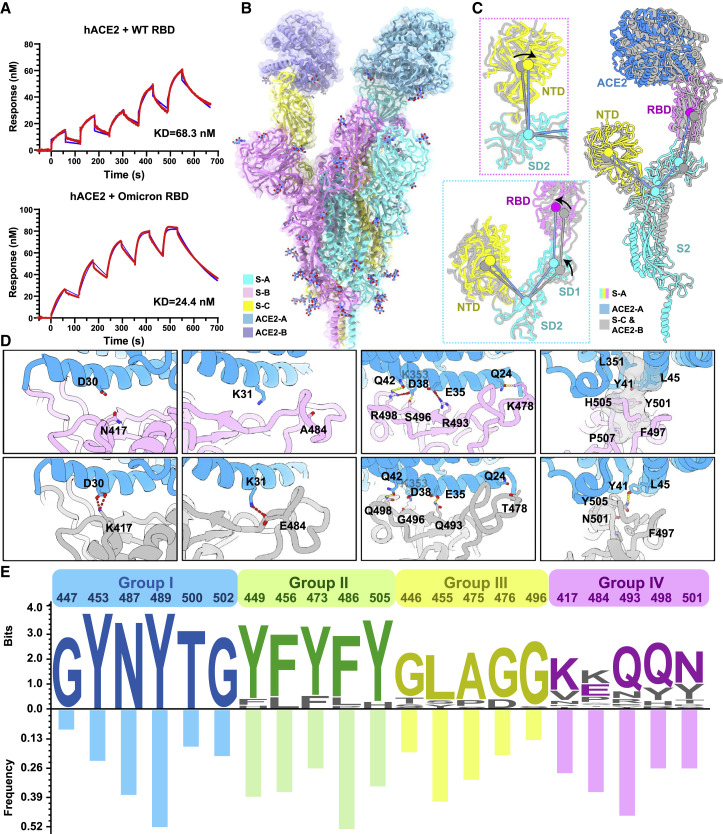
Figure 6.
Molecular determinants for enhanced binding affinity to human ACE2
(A) Binding affinity of hACE2 with WT RBD (top) or Omicron RBD (bottom) measured by SPR.
(B) Overall structure of the Omicron S-trimer in complex with hACE2. Three copies of S monomer were colored in yellow, cyan, and magenta, respectively; two hACE2 molecules bound to RBD were colored in purple and blue, respectively.
(C) Superimposition of two S monomer-hACE2 molecules with a focused alignment on S2 subunit. Color scheme for the S monomer in a stabilized up conformation is same as in Figure 1B, and the other S monomer is colored in gray. Insets represent the structural shifts of NTD and SD2 (left-top) and altered angles (left-bottom) formed by NTD, SD2, SD1, and RBD that were triggered by ACE2 binding.
(D) Changes at the interfaces between WT RBD (PDB: 6M0J) (Lan et al., 2020) and Omicron RBD with hACE2. hACE2, WT RBD, and Omicron RBD are colored in blue, gray, and magenta, respectively; key mutated residues were shown as sticks with the abolished (left four) and newly established (right four) bonds denoted in dashed lines. Salt bridges, red; hydron bonds, yellow. Hydrophobic network is highlighted in gray.
(E) Analysis of sequence conservation and antigenicity frequency on residues involved in ACE2 binding. The logo plot represents the conservation of these residues from 25 sarbecoviruses, and the histogram shows the antigenic frequency of the same residues targeted by NAbs. Logos and bars of four types of residues are colored with blue (identical residues), green (homologous residues), yellow (conditionally altered residues), and pink (highly diverse residues), respectively. Amino acid sequences of these residues involved in hACE2 binding are from WT SARS-CoV-2, 17 SARS-CoV-2 variants (Alpha, Beta, Gamma, Delta, Lambda, Mu, Delta plus, Omicron, Eta, Lota, Kappa, Theta, Iota, B.1.1.318, B.1.620, and C.1.2, C.363), SARS-CoV-1, pangolin coronavirus (GX/P2V/2017 and GD/1/2019), bat coronavirus (WIV, RaTG13 and LYRa11), and civet coronavirus of 007/2004.
See also Figures S1 and S6.