Figure 1.
The reprogramming efficiency of Nanog-GFP MEF using KLF4 ZnF alanine-scanned mutants
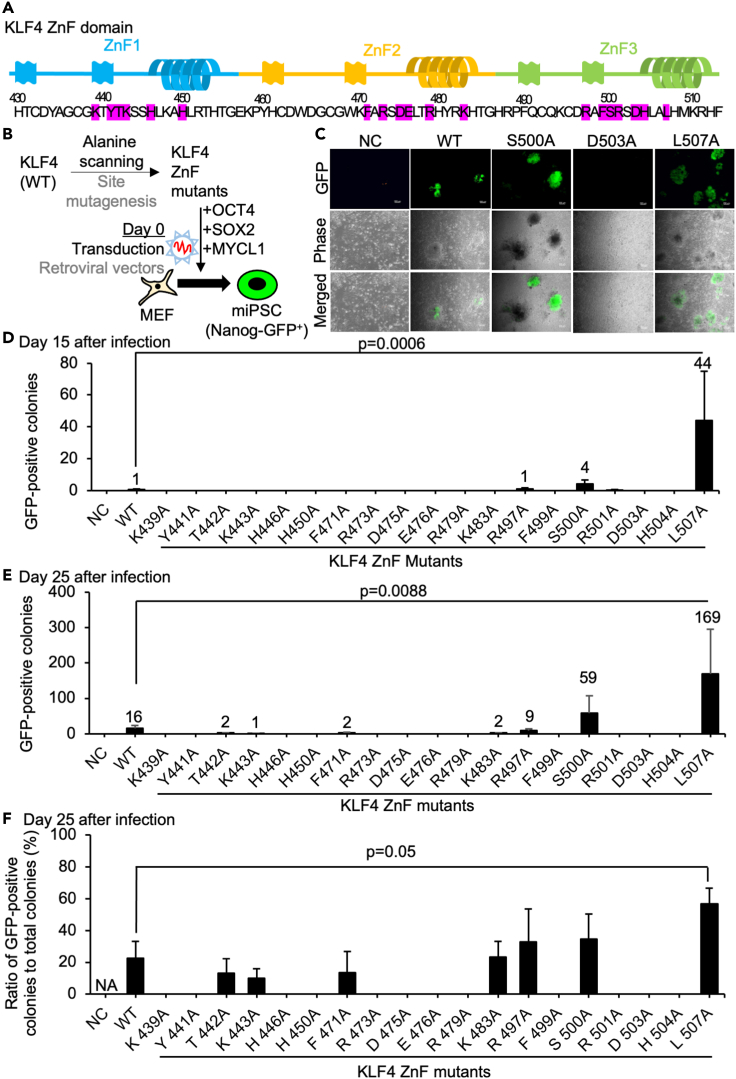
(A) Schematic of the protein sequence of the KLF4 ZnF domain. The amino acid residues highlighted in red are targeted by alanine scanning site mutagenesis in this study.
(B) Schematic of the generation of iPSCs using alanine-scanned KLF4 ZnF mutants made by site mutagenesis of KLF4 wild-type (WT) retroviral vector together with OCT4, SOX2, and MYCL1 retroviral vectors.
(C) The typical morphologies of generated iPSC colonies in NC (negative control with mRFP1 vector), WT (KLF4 wild-type), R497A mutant, D503A mutant, and L507A mutant. The images were shown in GFP (green channel fluorescence), Phase (phase contrast), and merged images. Scale bars, 200 μm.
(D) The number of Nanog-GFP-positive iPSC colonies were counted 15 days after infection of retroviral vectors to Nanog-GFP MEF from 10,000 cells. Results are mean and SE, n = 4. p value was calculated from Dunnett's test between WT and L507A conditions.
(E) The number of Nanog-GFP-positive colonies were counted 25 days after infection of retroviral vectors to Nanog-GFP MEF from 10,000 cells. Results are mean and SE, n = 4. p value was calculated from Dunnett's test between WT and L507A conditions.
(F) The ratio of Nanog-GFP-positive colonies was calculated 25 days after infection of retroviral vectors. The total number of Nanog-GFP-positives was counted from whole wells. Results are mean and SE, n = 4. p value was calculated from Dunnett's test between WT and L507A conditions.